Screening of SNP Loci Related to Leg Length Trait in Leizhou Goats Based on Whole-Genome Resequencing
- PMID: 39596516
- PMCID: PMC11594888
- DOI: 10.3390/ijms252212450
Screening of SNP Loci Related to Leg Length Trait in Leizhou Goats Based on Whole-Genome Resequencing
Abstract
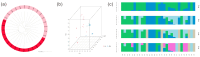
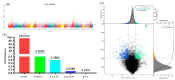
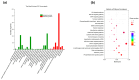
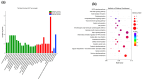
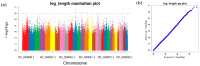
Leizhou goats can be classified into tall and short types based on their size and habits. The tall Leizhou goats are well-suited for grazing management due to their robust physique, while the dwarf types are smaller, grow rapidly, and are more appropriate for feeding management systems. In this study, whole-genome resequencing was conducted to identify genomic variants in 15 Tall-legged (TL) and 15 Short-legged (SL) Leizhou goats, yielding 8,641,229 high-quality SNPs in the Leizhou goat genome. Phylogenetic tree and principal component analyses revealed obvious genetic differentiation between the two groups. Fst and θπ analyses identified 420 genes in the TL group and 804 genes in the SL group. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses indicated that the phosphatidylinositol signaling system is associated with growth and development. Additionally, Genome-Wide Association Study (GWAS) analysis identified eight genes linked to leg length, including B4GALT7 and NR1D1. Notably, the NC_030818.1 (g.53666634T > C) variant was significantly associated with leg length traits, where the CC genotype was linked to shorter legs and the TT genotype to longer legs. This study identifies candidate genes and molecular markers, serving as a reference point for breeding and genetic improvement efforts in Leizhou goats and other goat breeds.
Keywords: Leizhou goat; SNPs; leg length trait; whole-genome resequencing.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Genome-wide association study of growth and reproductive traits based on low-coverage whole-genome sequencing in a Chubao black-head goat population.Gene. 2024 Dec 30;931:148891. doi: 10.1016/j.gene.2024.148891. Epub 2024 Aug 24. Gene. 2024. PMID: 39187139
-
Genome-wide association study for cashmere traits in Inner Mongolia cashmere goat population reveals new candidate genes and haplotypes.BMC Genomics. 2024 Jul 2;25(1):658. doi: 10.1186/s12864-024-10543-4. BMC Genomics. 2024. PMID: 38956486 Free PMC article.
-
Genome-wide association study identifies candidate genes affecting body conformation traits of Zhongwei goat.BMC Genomics. 2025 Jan 14;26(1):37. doi: 10.1186/s12864-024-11097-1. BMC Genomics. 2025. PMID: 39810085 Free PMC article.
-
Advances in genome-wide association studies for important traits in sheep and goats.Yi Chuan. 2017 Jun 20;39(6):491-500. doi: 10.16288/j.yczz.17-021. Yi Chuan. 2017. PMID: 28903908 Review.
-
Identifying Candidate Genes for Litter Size and Three Morphological Traits in Youzhou Dark Goats Based on Genome-Wide SNP Markers.Genes (Basel). 2023 May 29;14(6):1183. doi: 10.3390/genes14061183. Genes (Basel). 2023. PMID: 37372363 Free PMC article. Review.
Cited by
-
Examination of Runs of Homozygosity Distribution Patterns and Relevant Candidate Genes of Potential Economic Interest in Russian Goat Breeds Using Whole-Genome Sequencing.Genes (Basel). 2025 May 24;16(6):631. doi: 10.3390/genes16060631. Genes (Basel). 2025. PMID: 40565522 Free PMC article.
-
Leveraging Whole-Genome Resequencing to Uncover Genetic Diversity and Promote Conservation Strategies for Ruminants in Asia.Animals (Basel). 2025 Mar 13;15(6):831. doi: 10.3390/ani15060831. Animals (Basel). 2025. PMID: 40150358 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

