Genetic Diversity and Population Structure Analysis of Soybean [ Glycine max (L.) Merrill] Genotypes Using Agro-Morphological Traits and SNP Markers
- PMID: 39596572
- PMCID: PMC11593782
- DOI: 10.3390/genes15111373
Genetic Diversity and Population Structure Analysis of Soybean [ Glycine max (L.) Merrill] Genotypes Using Agro-Morphological Traits and SNP Markers
Abstract
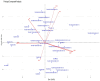
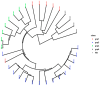
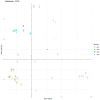
Background/Objectives: Understanding the genetic diversity of soybean genotypes can provide valuable information that guides parental selection and the design of an effective hybridization strategy in a soybean breeding program. In order to identify genetically diverse, complementary, and prospective parental lines for breeding, this study set out to ascertain the genetic diversity, relationships, and population structure among 35 soybean genotypes based on agro-morphological traits and Single Nucleotide Polymorphic (SNP) marker data. Methods/Results: Cluster analysis, based on agro-morphological traits, grouped the studied genotypes into four clusters. The first two principal components accounted for 62.8% of the total phenotypic variation, where days to 50% flowering, days to 95% maturity, grain yield, shattering score, and lodging score had high and positive contributions to the total variation. Using the SNP marker information, mean values of 0.16, 0.19, 0.067, and 0.227 were obtained for minor allele frequency (MAF), polymorphic information content (PIC), observed heterozygosity (Ho), and expected heterozygosity (He), respectively. Using different clustering approaches (admixture population structure, principal component scatter plot, and hierarchical clustering), the studied genotypes were grouped into four major clusters. Conclusions:The agro-morphological and molecular analysis results indicated the existence of moderate genetic diversity among the studied genotypes. The traits identified to be significantly related to yield provide valuable information for the genetic improvement of soybeans for yield.
Keywords: SNP Marker; genetic diversity; grain yield; soybean.
Conflict of interest statement
The authors declare there are no conflicts of interest.
Figures


References
-
- Chander S., Garcia-Oliveira A.L., Gedil M., Shah T., Otusanya G.O., Asiedu R., Chigeza G. Genetic diversity and population structure of soybean lines adapted to sub-Saharan Africa using single nucleotide polymorphism (SNP) markers. Agronomy. 2021;11:604. doi: 10.3390/agronomy11030604. - DOI
-
- Khojely D.M., Ibrahim S.E., Sapey E., Han T. History, current status, and prospects of soybean production and research in sub-Saharan Africa. Crop. J. 2018;6:226–235. doi: 10.1016/j.cj.2018.03.006. - DOI
-
- Lukanda M.M., Dramadri I.O., Adjei E.A., Arusei P., Gitonga H.W., Wasswa P., Edema R., Ssemakula M.O., Tukamuhabwa P., Tusiime G. Genetic Diversity and Population Structure of Ugandan Soybean (Glycine max L.) Germplasm Based on DArTseq. Plant Mol. Biol. Rep. 2023;41:417–426. doi: 10.1007/s11105-023-01375-9. - DOI
-
- Sivabharathi R., Muthuswamy A., Anandhi K., Karthiba L. Genetic Diversity Studies of Soybean [Glycine max (L.) Merrill] Germplasm Accessions using Cluster and Principal Component Analysis. Legume Res.—Int. J. 2023;1:6. doi: 10.18805/LR-5071. - DOI
-
- Abebe A.T., Kolawole A.O., Unachukwu N., Chigeza G., Tefera H., Gedil M. Assessment of diversity in tropical soybean (Glycine max (L.) Merr.) varieties and elite breeding lines using single nucleotide polymorphism markers. Plant Genet. Resour. 2021;19:20–28. doi: 10.1017/S1479262121000034. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

