Molecular and Functional Assessment of TSC1 and TSC2 in Individuals with Tuberous Sclerosis Complex
- PMID: 39596632
- PMCID: PMC11593644
- DOI: 10.3390/genes15111432
Molecular and Functional Assessment of TSC1 and TSC2 in Individuals with Tuberous Sclerosis Complex
Abstract
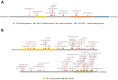
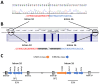
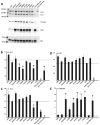
Tuberous sclerosis complex (TSC) is an autosomal dominant neurodevelopmental disorder and multisystem disease caused by pathogenic DNA alterations in the TSC1 and TSC2 tumor suppressor genes. A molecular genetic diagnosis of TSC confirms the clinical diagnosis, facilitating the implementation of appropriate care and surveillance. TSC1 and TSC2 encode the core components of the TSC1/2 complex (TSC1/2), a negative regulator of the mechanistic target of rapamycin (MTOR) complex 1 (TORC1). Functional analysis of the effects of TSC1 and TSC2 variants on TORC1 activity can help establish variant pathogenicity. We searched for pathogenic alterations to TSC1 and TSC2 in DNA isolated from 116 individuals with a definite clinical diagnosis of TSC. Missense variants and in-frame deletions were functionally assessed. Pathogenic DNA alterations were identified in 106 cases (91%); 18 (17%) in TSC1 and 88 (83%) in TSC2. Of these, 35 were novel. Disruption of TSC1/2 activity was demonstrated for seven TSC2 variants. Molecular diagnostics confirms the clinical diagnosis of TSC in a large proportion of cases. Functional assessment can help establish variant pathogenicity and is a useful adjunct to DNA analysis.
Keywords: Alu repeat; TSC1; TSC2; missense variant; neurodevelopmental disorder; pathogenic alteration; splicing variant; tuberous sclerosis complex.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Northrup H., Krueger D.A., International Tuberous Sclerosis Complex Consensus Group Tuberous sclerosis complex diagnostic criteria update: Recommendations of the 2012 International Tuberous Sclerosis Complex Consensus Conference. Pediatr. Neurol. 2013;49:243–254. doi: 10.1016/j.pediatrneurol.2013.08.001. - DOI - PMC - PubMed
-
- Northrup H., Aronow M.E., Bebin E.M., Bissler J., Darling T.N., de Vries P.J., Frost M.D., Fuchs Z., Gosnell E.S., Gupta N., et al. Updated International Tuberous Sclerosis Complex Diagnostic Criteria and Surveillance and Management Recommendations. Pediatr. Neurol. 2021;123:50–66. doi: 10.1016/j.pediatrneurol.2021.07.011. - DOI - PubMed
-
- Au K.S., Williams A.T., Roach E.S., Batchelor L., Sparagana S.P., Delgado M.R., Wheless J.W., Baumgartner J.E., Roa B.B., Wilson C.M., et al. Genotype/phenotype correlation in 325 individuals referred for a diagnosis of tuberous sclerosis complex in the United States. Genet. Med. 2007;9:88–100. doi: 10.1097/GIM.0b013e31803068c7. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 1570747 and 88881.132401/2016-01/Coordenação de Aperfeicoamento de Pessoal de Nível Superior
- 2011/14329-9, 2013/08028-1 and 2019/10868-4/Fundação de Amparo à Pesquisa do Estado de São Paulo
- 2016-P07/Tuberous Sclerosis Association
- Process 20380009/2024/Brazil Ministry of Education
- 00/Michelle Foundation (The Netherlands)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

