Structural Equation Modelling as a Proof-of-Concept Tool for Mediation Mechanisms Between Topical Antibiotic Prophylaxis and Six Types of Blood Stream Infection Among ICU Patients
- PMID: 39596789
- PMCID: PMC11591272
- DOI: 10.3390/antibiotics13111096
Structural Equation Modelling as a Proof-of-Concept Tool for Mediation Mechanisms Between Topical Antibiotic Prophylaxis and Six Types of Blood Stream Infection Among ICU Patients
Abstract
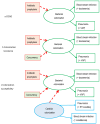
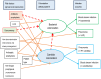
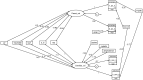
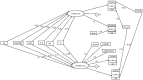
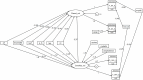
Whether exposing the microbiome to antibiotics decreases or increases the risk of blood stream infection with Pseudomonas aeruginosa, Staphylococcus aureus, Acinetobacter, and Candida among ICU patients, and how this altered risk might be mediated, are critical research questions. Addressing these questions through the direct study of specific constituents within the microbiome would be difficult. An alternative tool for addressing these research questions is structural equation modelling (SEM). SEM enables competing theoretical causation networks to be tested 'en bloc' by confrontation with data derived from the literature. These causation models have three conceptual steps: exposure to specific antimicrobials are the key drivers, clinically relevant infection end points are the measurable observables, and the activity of key microbiome constituents on microbial invasion serve as mediators. These mediators, whether serving to promote, to impede, or neither, are typically unobservable and appear as latent variables in each model. SEM methods enable comparisons through confronting the three competing models, each versus clinically derived data with the various exposures, such as topical or parenteral antibiotic prophylaxis, factorized in each model. Candida colonization, represented as a latent variable, and concurrency are consistent promoters of all types of blood stream infection, and emerge as harmful mediators.
Keywords: Acinetobacter; Candida; Pseudomonas aeruginosa; Staphylococcus aureus; bacteremia; gut/blood microbiome; intensive care; structural equation modelling.
Conflict of interest statement
The author declares no conflicts of interest.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

