Differential Host Gene Expression in Response to Infection by Different Mycobacterium tuberculosis Strains-A Pilot Study
- PMID: 39597535
- PMCID: PMC11596623
- DOI: 10.3390/microorganisms12112146
Differential Host Gene Expression in Response to Infection by Different Mycobacterium tuberculosis Strains-A Pilot Study
Abstract
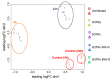
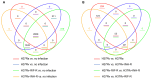
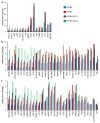
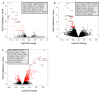
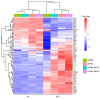
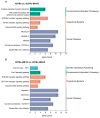
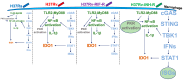
Tuberculosis (TB) represents a global public health threat and is a leading cause of morbidity and mortality worldwide. Effective control of TB is complicated with the emergence of multidrug resistance. Yet, there is a fundamental gap in understanding the complex and dynamic interactions between different Mycobacterium tuberculosis strains and the host. In this pilot study, we investigated the host immune response to different M. tuberculosis strains, including drug-sensitive avirulent or virulent, and rifampin-resistant or isoniazid-resistant virulent strains in human THP-1 cells. We identified major differences in the gene expression profiles in response to infection with these strains. The expression of IDO1 and IL-1β in the infected cells was stronger in all virulent M. tuberculosis strains. The most striking result was the overexpression of many interferon-stimulated genes (ISGs) in cells infected with the isoniazid-resistant strain, compared to the rifampin-resistant and the drug-sensitive strains. Our data indicate that infection with the isoniazid-resistant M. tuberculosis strain preferentially resulted in cGAS-STING/STAT1 activation, which induced a characteristic host immune response. These findings reveal complex gene signatures and a dynamic variation in the immune response to infection by different M. tuberculosis strains.
Keywords: Mycobacterium tuberculosis; antimicrobial resistance; expression profiling genes; host–pathogen interaction; interferon-stimulated genes (ISGs).
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- World Health Organization (WHO) Global Tuberculosis Report 2022. [(accessed on 22 June 2023)]. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/globa....
-
- Chakaya J., Petersen E., Nantanda R., Mungai B.N., Migliori G.B., Amanullah F., Lungu P., Ntoumi F., Kumarasamy N., Maeurer M., et al. The WHO global tuberculosis 2021 report—Not so good news and turning the tide back to end TB. Int. J. Infect. Dis. 2022;124:S26–S29. doi: 10.1016/j.ijid.2022.03.011. - DOI - PMC - PubMed
-
- Klopper M., Warren R.M., Hayes C., van Pittius N.C.G., Streicher E.M., Müller B., Sirgel F.A., Chabula-Nxiweni M., Hoosain E., Coetzee G., et al. Emergence and spread of extensively and totally drug-resistant tuberculosis, South Africa. Emerg. Infect. Dis. 2013;19:449–455. doi: 10.3201/eid1903.120246. - DOI - PMC - PubMed
-
- Zhan L., Wang J., Wang L., Qin C. The correlation of drug resistance and virulence in Mycobacterium tuberculosis. Biosaf. Health. 2020;2:18–24. doi: 10.1016/j.bsheal.2020.02.004. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

