Exiguolysin, a Novel Thermolysin (M4) Peptidase from Exiguobacterium oxidotolerans
- PMID: 39597700
- PMCID: PMC11596557
- DOI: 10.3390/microorganisms12112311
Exiguolysin, a Novel Thermolysin (M4) Peptidase from Exiguobacterium oxidotolerans
Abstract
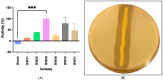
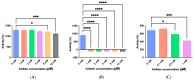
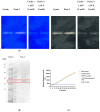
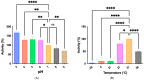
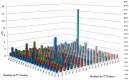
This study details a comprehensive biochemical and structural characterization of exiguolysin, a novel thermolysin-like, caseinolytic peptidase secreted by a marine isolate of Exiguobacterium oxidotolerans strain BW26. Exiguolysin demonstrated optimal proteolytic activity at 37 °C and pH 3, retaining 85% activity at 50 °C, highlighting its potential stability under broad reaction conditions. SDS-PAGE and LC-MS analysis identified the enzyme as a 32 kDa M4-family metalloprotease. Exiguolysin activity was inhibited by 1,10-phenanthroline, confirming its dependence on metal ions for activity. Zymographic analysis and substrate specificity assays revealed selective hydrolysis of matrix metalloproteinase (MMP) substrates but no activity against elastase substrates. Analysis of the predicted gene sequence and structural predictions using AlphaFold identified the presence and position of HEXXH and Glu-Xaa-Xaa-Xaa-Asp motifs, crucial for zinc binding and catalytic activity, characteristic of 'Glu-zincins' and members of the M4 peptidase family. High-throughput screening of a 20 × 20 N-alpha mercaptoamide dipeptide inhibitor library against exiguolysin identified SH-CH2-CO-Met-Tyr-NH2 as the most potent inhibitor, with a Ki of 1.95 μM. Notably, exiguolysin selectively inhibited thrombin-induced PAR-1 activation in PC-3 cells, potentially indicating a potential mechanism of virulence in modulating PAR-1 signalling during infection by disarming PARs. This is the first detailed characterization of a peptidase of the M4 (thermolysin) family in the genus Exiguobacterium which may have industrial application potential and relevance as a putative virulence factor.
Keywords: Exiguobacterium oxidotolerans; metalloprotease; protease-activated receptors; thermolysin.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Collins M., Lund B., Farrow J., Schleifer K. Chemotaxonomic study of an alkalophilic bacterium, Exiguobacterium aurantiacum gen. nov., sp. nov. Microbiology. 1983;129:2037–2042. doi: 10.1099/00221287-129-7-2037. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

