SARS-CoV-2 Genomic Variants and Their Relationship with the Expressional and Genomic Profile of Angiotensin-Converting Enzyme 2 (ACE2) and Transmembrane Serine Protease 2 (TMPRSS2)
- PMID: 39597701
- PMCID: PMC11596641
- DOI: 10.3390/microorganisms12112312
SARS-CoV-2 Genomic Variants and Their Relationship with the Expressional and Genomic Profile of Angiotensin-Converting Enzyme 2 (ACE2) and Transmembrane Serine Protease 2 (TMPRSS2)
Abstract
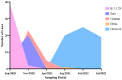
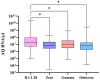
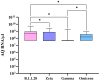
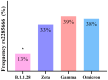
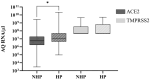
Over the past four years, angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) have been extensively studied, given their important role in SARS-CoV-2 replication; however, most studies have failed to compare their behavior in the face of different SARS-CoV-2 genomic variants. Therefore, this study evaluated the influence of different variants in ACE2/TMPRSS2 expressional and genomic profiles. To achieve this, 160 nasopharyngeal samples, previously detected with SARS-CoV-2 via RT-qPCR (June 2020-July 2022), were quantified for ACE2/TMPRSS2 expression levels, also using RT-qPCR; SARS-CoV-2 genomic variants, along with polymorphisms in the ACE2/TMPRSS2 coding genes, were identified using nanopore sequencing. In order of appearance, the B.1.1.28, Zeta, Gamma, and Omicron variants were identified in this study. The ACE2 levels were higher when B.1.1.28 was present, possibly due to the ACE2/spike binding affinity; the TMPRSS2 levels were also higher in the presence of B.1.1.28, probably attributable to inefficient usage of the TMPRSS2 pathway by the other variants, as well as to the decrease in protease transcription factors when in the presence of Omicron. The rs2285666 (ACE2) polymorphism was less frequent when B.1.1.28 was present, which is befitting, since rs2285666 increases ACE2/spike binding affinity. In conclusion, SARS-CoV-2 genomic variants appear to exhibit varying impacts in regards to ACE2/TMPRSS2 genomic and expressional behavior.
Keywords: genomic variants; host–parasite; viral infection.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., Doddato G., Giliberti A., Marconi C., Musacchia F., et al. ACE2 Gene Variants May Underlie Interindividual Variability and Susceptibility to COVID-19 in the Italian Population. Eur. J. Hum. Genet. 2020;28:1602–1614. doi: 10.1038/s41431-020-0691-z. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

