Differential Cytotoxic Effects of Cell-Free Supernatants of Emerging Pathogens Escherichia albertii and Escherichia fergusonii on Four Cell Lines Reveal Vero Cells as a Putative Candidate for Cytotoxicity Analysis
- PMID: 39597758
- PMCID: PMC11596466
- DOI: 10.3390/microorganisms12112370
Differential Cytotoxic Effects of Cell-Free Supernatants of Emerging Pathogens Escherichia albertii and Escherichia fergusonii on Four Cell Lines Reveal Vero Cells as a Putative Candidate for Cytotoxicity Analysis
Abstract
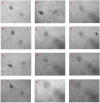
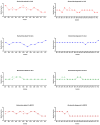
Escherichia albertii and Escherichia fergusonii are recognized as emerging pathogens with zoonotic potential. Despite their increasing importance, there is a paucity of data on the cytotoxicity of these two pathogens. Therefore, in the present study, we investigated the cytotoxic potentials of the cell-free supernatants from 10 E. albertii and 15 E. fergusonii isolates for their cytotoxic effects on four different cell lines (CHO, Vero, HeLa, and MDCK). All E. albertii isolates (100%) and all but one E. fergusonii (93.33%) were cytotoxic. E. albertii isolates produced similar cytotoxicity titres across the cell lines, whereas the Vero cell was found to be the most sensitive to toxins produced by E. fergusonii (p < 0.05), followed by HeLa and CHO cells. MDCK was the least sensitive cell line to E. fergusonii toxins (p < 0.05). PCR detection of cytotoxicity-associated genes (cdtB, stx1, and stx2) indicated uniform possession of cdtB gene by all E. albertii isolates, while stx1 and stx2 genes were harboured neither by E. albertii, nor E. fergusonii. Taken together, our results provided experimental evidence of the cytotoxic effects of these two emerging pathogens, and Vero cells were identified as an optimal candidate to study the cytotoxic effects of E. albertii and E. fergusonii.
Keywords: CDT; Escherichia albertii; Escherichia fergusonii; HeLa cells; Vero cells; cytotoxicity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Romão F.T., Martins F.H., Hernandes R.T., Ooka T., Santos F.F., Yamamoto D., Bonfim-Melo A., Jones N., Hayashi T., Elias W.P., et al. Genomic Properties and Temporal Analysis of the Interaction of an Invasive Escherichia albertii With Epithelial Cells. Front. Cell. Infect. Microbiol. 2020;10:571088. doi: 10.3389/fcimb.2020.571088. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

