Genomic Insights of Wheat Root-Associated Lysinibacillus fusiformis Reveal Its Related Functional Traits for Bioremediation of Soil Contaminated with Petroleum Products
- PMID: 39597765
- PMCID: PMC11596681
- DOI: 10.3390/microorganisms12112377
Genomic Insights of Wheat Root-Associated Lysinibacillus fusiformis Reveal Its Related Functional Traits for Bioremediation of Soil Contaminated with Petroleum Products
Abstract
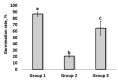
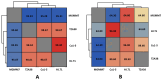
The negative ecological impact of industrialization, which involves the use of petroleum products and dyes in the environment, has prompted research into effective, sustainable, and economically beneficial green technologies. For green remediation primarily based on active microbial metabolites, these microbes are typically from relevant sources. Active microbial metabolite production and genetic systems involved in xenobiotic degradation provide these microbes with the advantage of survival and proliferation in polluted ecological niches. In this study, we evaluated the ability of wheat root-associated L. fusiformis MGMM7 to degrade xenobiotic contaminants such as crude oil, phenol, and azo dyes. We sequenced the whole genome of MGMM7 and provided insights into the genomic structure of related strains isolated from contaminated sources. The results revealed that influenced by its isolation source, L. fusiformis MGMM7 demonstrated remediation and plant growth-promoting abilities in soil polluted with crude oil. Lysinibacillus fusiformis MGMM7 degraded up to 44.55 ± 5.47% crude oil and reduced its toxicity in contaminated soil experiments with garden cress (Lepidium sativum L.). Additionally, L. fusiformis MGMM7 demonstrated a significant ability to degrade Congo Red azo dye (200 mg/L), reducing its concentration by over 60% under both static and shaking cultivation conditions. However, the highest degradation efficiency was observed under shaking conditions. Genomic comparison among L. fusiformis strains revealed almost identical genomic profiles associated with xenobiotic assimilation. Genomic relatedness using Average Nucleotide Identity (ANI) and digital DNA-DNA hybridization (DDH) revealed that MGMM7 is distantly related to TZA38, Cu-15, and HJ.T1. Furthermore, subsystem distribution and pangenome analysis emphasized the distinctive features of MGMM7, including functional genes in its chromosome and plasmid, as well as the presence of unique genes involved in PAH assimilation, such as phnC/T/E, which is involved in phosphonate biodegradation, and nemA, which is involved in benzoate degradation and reductive degradation of N-ethylmaleimide. These findings highlight the potential properties of petroleum-degrading microorganisms isolated from non-contaminated rhizospheres and offer genomic insights into their functional diversity for xenobiotic remediation.
Keywords: azo dye; crude oil; pangenome; phenol; polycyclic aromatic hydrocarbons; xenobiotic assimilation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








References
-
- Ajibade F.O., Adelodun B., Ajibade T.F., Lasisi K.H., Abiola C., Adewumi J.R., Akinbile C.O. The threatening effects of open dumping on soil at waste disposal sites of Akure city, Nigeria. Int. J. Environ. Waste Manag. 2021;27:127–146. doi: 10.1504/IJEWM.2021.112947. - DOI
-
- Hussain B., Chen J.S., Huang S.W., Tsai I.S., Rathod J., Hsu B.M. Underpinning the ecological response of mixed chlorinated volatile organic compounds (CVOCs) associated with contaminated and bioremediated groundwaters: A potential nexus of microbial community structure and function for strategizing efficient bioremediation. Environ. Pollut. 2023;334:122–215. doi: 10.1016/j.envpol.2023.122215. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

