Identification of Estrogen-Responsive Proteins in Mouse Seminal Vesicles Through Mass Spectrometry-Based Proteomics
- PMID: 39598420
- PMCID: PMC11597337
- DOI: 10.3390/ph17111508
Identification of Estrogen-Responsive Proteins in Mouse Seminal Vesicles Through Mass Spectrometry-Based Proteomics
Abstract
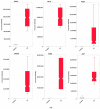
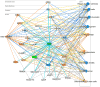
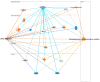
Background: Although estrogenic compounds promise therapeutic potential in treating various conditions, concerns regarding their endocrine-disrupting effects have been raised. Current methodologies for screening estrogenicity in rodent models are limited to the female-specific uterotrophic bioassay. Studies have reported enlargement of the seminal vesicles in orchiectomized males treated with estrogens. However, identifying estrogenicity strictly through changes in wet weights is uninformative regarding the molecular mechanisms of these agents. Therefore, protein-based biomarkers can complement and improve the sensitivity of weight-based assessments. To this end, we present a discovery-driven proteomic analysis of 17β-estradiol's effects on the seminal vesicles. Methods: We treated orchidectomized mice with the hormone for five days and used the vehicle-treated group as a control. Seminal vesicles were analyzed by shotgun approach using data-dependent nanoflow liquid chromatography-tandem mass spectrometry and label-free quantification. Proteins found to be differentially expressed between the two groups were processed through a bioinformatics pipeline focusing on pathway analyses and assembly of protein interaction networks. Results: Out of 668 identified proteins that passed rigorous validation criteria, 133 were regulated significantly by 17β-estradiol. Ingenuity Pathway Analysis® linked them to several hormone-affected pathways, including those associated with immune function such as neutrophil degranulation. The altered protein interaction networks were also related to functions including endocrine disruption, abnormal metabolism, and therapeutic effects. Conclusions: We identified several potential biomarkers for estrogenicity in mouse seminal vesicles, many of them not previously linked with exogenous 17β-estradiol exposure.
Keywords: 17β-estradiol; bioinformatics; biomarker discovery; endocrine disruption; male reproductive system; mouse seminal vesicle; proteomics; therapeutic effects.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Proteomics Complementation of the Rat Uterotrophic Assay for Estrogenic Endocrine Disruptors: A Roadmap of Advancing High Resolution Mass Spectrometry-Based Shotgun Survey to Targeted Biomarker Quantifications.Int J Mol Sci. 2021 Feb 8;22(4):1686. doi: 10.3390/ijms22041686. Int J Mol Sci. 2021. PMID: 33567512 Free PMC article.
-
Sensitivity and specificity of the bioassay of estrogenicity in mammary gland and seminal vesicles of male mice.Physiol Res. 2002;51(3):267-76. Physiol Res. 2002. PMID: 12234119
-
Molecular feminization of mouse seminal vesicle by prenatal exposure to diethylstilbestrol: altered expression of messenger RNA.J Urol. 1994 May;151(5):1370-8. doi: 10.1016/s0022-5347(17)35263-1. J Urol. 1994. PMID: 8158792
-
Critical review and evaluation of the uterotrophic bioassay for the identification of possible estrogen agonists and antagonists: in support of the validation of the OECD uterotrophic protocols for the laboratory rodent. Organisation for Economic Co-operation and Development.Crit Rev Toxicol. 2002;32(6):445-520. doi: 10.1080/20024091064291. Crit Rev Toxicol. 2002. PMID: 12487363 Review.
-
Estrogenic Endocrine Disrupting Chemicals Influencing NRF1 Regulated Gene Networks in the Development of Complex Human Brain Diseases.Int J Mol Sci. 2016 Dec 13;17(12):2086. doi: 10.3390/ijms17122086. Int J Mol Sci. 2016. PMID: 27983596 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

