Interrelation Between Pathoadaptability Factors and Crispr-Element Patterns in the Genomes of Escherichia coli Isolates Collected from Healthy Puerperant Women in Ural Region, Russia
- PMID: 39599550
- PMCID: PMC11597047
- DOI: 10.3390/pathogens13110997
Interrelation Between Pathoadaptability Factors and Crispr-Element Patterns in the Genomes of Escherichia coli Isolates Collected from Healthy Puerperant Women in Ural Region, Russia
Abstract
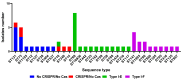
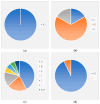
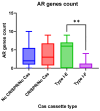
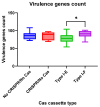
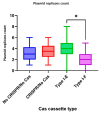
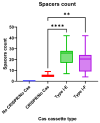
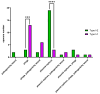
Escherichia coli is a commensal and opportunistic bacterium widely distributed around the world in different niches including intestinal of humans and animals, and its extraordinary genome plasticity led to the emergence of pathogenic strains causing a wide range of diseases. E. coli is one of the monitored species in maternity hospitals, being the main etiological agent of urogenital infections, endometriosis, puerperal sepsis, and neonatal diseases. This study presents a comprehensive analysis of E. coli isolates obtained from the maternal birth canal of healthy puerperant women 3-4 days after labor. According to whole genome sequencing data, 31 sequence types and six phylogenetic groups characterized the collection containing 53 isolates. The majority of the isolates belonged to the B2 phylogroup. The data also includes phenotypic and genotypic antibiotic resistance profiles, virulence factors, and plasmid replicons. Phenotypic and genotypic antibiotic resistance testing did not demonstrate extensive drug resistance traits except for two multidrug-resistant E. coli isolates. The pathogenic factors revealed in silico were assessed with respect to CRISPR-element patterns. Multiparametric and correlation analyses were conducted to study the interrelation of different pathoadaptability factors, including antimicrobial resistance and virulence genomic determinants carried by the isolates under investigation. The data presented will serve as a valuable addition to further scientific investigations in the field of bacterial pathoadaptability, especially in studying the role of CRISPR/Cas systems in the E. coli genome plasticity and evolution.
Keywords: CRISPR/Cas system; Escherichia coli; WGS; antibiotic resistance genes; pathoadaptability; pathogenic potential; virulence factors.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Whole-genome sequencing-based phylogeny, antibiotic resistance, and invasive phenotype of Escherichia coli strains colonizing the cervix of women in preterm labor.BMC Microbiol. 2021 Dec 3;21(1):330. doi: 10.1186/s12866-021-02389-7. BMC Microbiol. 2021. PMID: 34861816 Free PMC article.
-
Molecular characterization of multidrug-resistant E. coli recovered from diarrheagenic children under 5 years from Mukuru Informal Settlement, Nairobi, Kenya, based on whole-genome sequencing analysis.Microbiol Spectr. 2025 Jun 3;13(6):e0142024. doi: 10.1128/spectrum.01420-24. Epub 2025 May 15. Microbiol Spectr. 2025. PMID: 40372033 Free PMC article.
-
Whole genome sequences of multi-drug resistant Escherichia coli isolated in a Pastoralist Community of Western Uganda: Phylogenomic changes, virulence and resistant genes.PLoS One. 2020 May 29;15(5):e0231852. doi: 10.1371/journal.pone.0231852. eCollection 2020. PLoS One. 2020. PMID: 32469885 Free PMC article.
-
Whole genome analysis of multidrug-resistant Escherichia coli isolate collected from drinking water in Armenia revealed the plasmid-borne mcr-1.1-mediated colistin resistance.Microbiol Spectr. 2024 Oct 3;12(10):e0075124. doi: 10.1128/spectrum.00751-24. Epub 2024 Aug 21. Microbiol Spectr. 2024. PMID: 39166856 Free PMC article.
-
[Population genetics of enterohemorrhagic Escherichia coli using whole-genome sequencing analyses].Nihon Saikingaku Zasshi. 2024;79(4):283-289. doi: 10.3412/jsb.79.283. Nihon Saikingaku Zasshi. 2024. PMID: 39631880 Review. Japanese.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

