Coxsackievirus A6 U.K. Genetic and Clinical Epidemiology Pre- and Post-SARS-CoV-2 Emergence
- PMID: 39599573
- PMCID: PMC11597771
- DOI: 10.3390/pathogens13111020
Coxsackievirus A6 U.K. Genetic and Clinical Epidemiology Pre- and Post-SARS-CoV-2 Emergence
Abstract
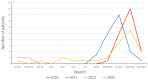
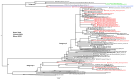
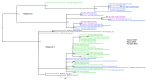
Coxsackievirus A6 (CVA6) has become increasingly clinically relevant as a cause of Hand, Foot and Mouth Disease (HFMD) globally since 2008. However, most laboratories do not routinely determine the enteroviral type of positive samples. The non-pharmaceutical measures introduced to curb transmission during the COVID-19 pandemic may also have perturbed CVA6 epidemiology. We thus aimed to determine the prevalence, clinical presentation and genetic relationship of CVA6 across three complete epidemic seasons: one pre-SARS-CoV-2 emergence and two post-SARS-CoV-2 emergence in our regional healthcare setting. Surplus diagnostic nucleic acid from diagnosed enteroviral positives diagnosed between September and December of 2018 and between May 2021 and April of 2023 was subject to VP1 gene sequencing to determine the CVA6 cases and interrogate their phylogenetic relationship. The confirmed CVA6 cases were also retrospectively clinically audited. CVA6 infections were identified in 33 and 69 individuals pre- and post-pandemic, respectively, with cases peaking in November of 2018 and 2022, but in October of 2021. HFMD was the primary diagnosis in 85.5% of the post-pandemic cases, but only 69.7% of the pre-pandemic cases, where respiratory and neurological symptoms (45.5% and 12.1%, respectively) were significantly elevated. A complete VP1 sequence was retrieved for 94% of the CVA6 cases, revealing that studied infections were genetically diverse and suggestive of multiple local and international transmission chains. CVA6 presented a significant clinical burden in our regional U.K. hospital setting both pre- and post-pandemic and was subject to dynamic clinical and genetic epidemiology.
Keywords: CVA6; HFMD; coxsackievirus A6; enterovirus; genetic epidemiology; hand, foot and mouth disease.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Prevalence of Coxsackievirus A6 and Enterovirus 71 in Hand, Foot and Mouth Disease in Nanjing, China in 2013.Pediatr Infect Dis J. 2015 Sep;34(9):951-7. doi: 10.1097/INF.0000000000000794. Pediatr Infect Dis J. 2015. PMID: 26090576
-
[Molecular epidemiology of HFMD-associated pathogen coxsackievirus A6 in Fujian Province, 2011-2013].Bing Du Xue Bao. 2014 Nov;30(6):624-9. Bing Du Xue Bao. 2014. PMID: 25868276 Chinese.
-
Clinical features of hand, foot and mouth disease caused by Coxsackievirus A6 in Xi'an, China, 2013-2019: A multicenter observational study.Acta Trop. 2024 Sep;257:107310. doi: 10.1016/j.actatropica.2024.107310. Epub 2024 Jun 30. Acta Trop. 2024. PMID: 38955319
-
Coxsackievirus A6 associated hand, foot and mouth disease in adults: clinical presentation and review of the literature.J Clin Virol. 2014 Aug;60(4):381-6. doi: 10.1016/j.jcv.2014.04.023. Epub 2014 May 9. J Clin Virol. 2014. PMID: 24932735 Review.
-
Seroprevalence and Virologic Surveillance of Enterovirus 71 and Coxsackievirus A6, United Kingdom, 2006-2017.Emerg Infect Dis. 2021 Sep;27(9):2261-2268. doi: 10.3201/eid2709.204915. Emerg Infect Dis. 2021. PMID: 34423767 Free PMC article. Review.
Cited by
-
Evaluating enterovirus diversity among symptomatic patients in Hungary during and after easing the COVID-19 lockdown.Virol J. 2025 Jun 24;22(1):204. doi: 10.1186/s12985-025-02835-2. Virol J. 2025. PMID: 40555984 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

