SMC5/6-Mediated Transcriptional Regulation of Hepatitis B Virus and Its Therapeutic Potential
- PMID: 39599784
- PMCID: PMC11598903
- DOI: 10.3390/v16111667
SMC5/6-Mediated Transcriptional Regulation of Hepatitis B Virus and Its Therapeutic Potential
Abstract
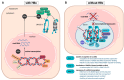
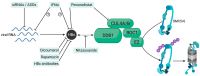
Cells have developed various mechanisms to counteract viral infections. In an evolutionary arms race, cells mobilize cellular restriction factors to fight off viruses, targeted by viral factors to facilitate their own replication. The hepatitis B virus (HBV) is a small dsDNA virus that causes acute and chronic infections of the liver. Its genome persists in the nuclei of infected hepatocytes as a covalently closed circular DNA (cccDNA) minichromosome, thus building up an episomal persistence reservoir. The chromosomal maintenance complex SMC5/6 acts as a restriction factor hindering cccDNA transcription, whereas the viral regulatory protein HBx targets SMC5/6 for proteasomal degradation, thus relieving transcriptional suppression of the HBV minichromosome. To date, no curative therapies are available for chronic HBV carriers. Knowledge of the factors regulating the cccDNA and the development of therapies involving silencing the minichromosome or specifically interfering with the HBx-SMC5/6 axis holds promise in achieving sustained viral control. Here, we summarize the current knowledge of the mechanism of SMC5/6-mediated HBV restriction. We also give an overview of SMC5/6 cellular functions and how this compares to the restriction of other DNA viruses. We further discuss the therapeutic potential of available and investigational drugs interfering with the HBx-SMC5/6 axis.
Keywords: HBx; NSE; SMC5/6; antiviral therapy; cccDNA; chronic hepatitis B; hepatitis B virus; interferon; siRNA; transcription.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- WHO Fact Sheet. [(accessed on 2 September 2024)]. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b.
-
- Locatelli M., Quivy J.P., Chapus F., Michelet M., Fresquet J., Maadadi S., Aberkane A.N., Diederichs A., Lucifora J., Rivoire M., et al. HIRA Supports Hepatitis B Virus Minichromosome Establishment and Transcriptional Activity in Infected Hepatocytes. Cell Mol. Gastroenterol. Hepatol. 2022;14:527–551. doi: 10.1016/j.jcmgh.2022.05.007. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

