Epidemiologic Investigation and Genetic Variation Analysis of PRRSV, PCV2, and PCV3 in Guangdong Province, China from 2020 to 2022
- PMID: 39599802
- PMCID: PMC11598979
- DOI: 10.3390/v16111687
Epidemiologic Investigation and Genetic Variation Analysis of PRRSV, PCV2, and PCV3 in Guangdong Province, China from 2020 to 2022
Abstract
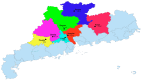
Recently, the emergence of HP-PRRSV (Highly Pathogenic porcine reproductive and respiratory syndrome virus) and the exacerbation of mixed infections of PRRSV and PCV have resulted in significant economic losses for the Chinese pig industry. This study collected a total of 226 samples suspected of infection with the aforementioned viruses from diverse pig farms in seven urban districts of central and northern Guangdong Province between 2020 and 2022. The positive rates of PRRSV, PCV2, and PCV3 in the samples were 33.2%, 37.6%, and 7.5%, respectively, and there were various mixed-infection scenarios present in the samples. This study successfully isolated multiple strains of PRRSV2 and PCV2 from their positive samples, and obtained the gene sequences of six PCV3 (ORF1 + ORF2) from samples. The associated sequences obtained were subjected to bioinformatic analysis and revealed the following:Predominantly prevalent strains of PRRSV in Guangdong Province include HP-PRRSV and NADC30-like variants, whereas PCV2 is primarily represented by the 2b and 2d subtypes. Specifically, the amino acid variation patterns exhibited by the PRRSV GP5 and NSP2 proteins of the strains sg_2108, qy_2008, and fs_2108 under environmental selective pressure are remarkably similar to the characteristics of Highly Pathogenic PRRSV; thus, it is inferred that they may possess higher virulence. The detected PCV3 strains were predominantly concentrated within the PCV3a-IM branch. All PRRSV strains involved in this study are wild-type-PRRSV (wt-PRRSV), comprising three recombinant strains and seven highly virulent strains. Among these strains, the ORF1a gene exhibited the highest variability in their genomes. Environmental selective pressure may enhance the virulence and immune evasion capabilities of PRRSV and drive mutations in the Cap proteins of PCV2 and PCV3. Conversely, PCV2 and PCV3 strains demonstrated greater stability in genetic evolution. In conclusion, this study enhances the epidemiological data regarding PRRSV, PCV2, and PCV3 in Guangdong Province, China, and is significant for the surveillance, prevention, and active control of these three diseases.
Keywords: epidemiological investigation; genetic variation analysis; porcine circovirus type 2 (PCV2); porcine circovirus type 3 (PCV3); porcine reproductive and respiratory syndrome virus (PRRSV).
Conflict of interest statement
Dongfang Zhao and Lianxiang Wang were employed by Wen’s Group Academy, Wen’s Foodstuffs Group Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures





Similar articles
-
Co-infection status of classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circoviruses (PCV2 and PCV3) in eight regions of China from 2016 to 2018.Infect Genet Evol. 2019 Mar;68:127-135. doi: 10.1016/j.meegid.2018.12.011. Epub 2018 Dec 17. Infect Genet Evol. 2019. PMID: 30572028
-
A novel NADC30-like porcine reproductive and respiratory syndrome virus (PRRSV) plays a limited role in the pathogenicity of porcine circoviruses (PCV2 and PCV3) and PRRSV co-infection.Transbound Emerg Dis. 2019 Jan;66(1):28-34. doi: 10.1111/tbed.13026. Epub 2018 Oct 13. Transbound Emerg Dis. 2019. PMID: 30267610
-
Prevalence and genetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) between 2018 and 2020 in central China.Infect Genet Evol. 2021 Oct;94:105016. doi: 10.1016/j.meegid.2021.105016. Epub 2021 Jul 27. Infect Genet Evol. 2021. PMID: 34325052
-
Novel Porcine Circoviruses in View of Lessons Learned from Porcine Circovirus Type 2-Epidemiology and Threat to Pigs and Other Species.Viruses. 2022 Jan 27;14(2):261. doi: 10.3390/v14020261. Viruses. 2022. PMID: 35215854 Free PMC article. Review.
-
Emergence, prevalence and evolution of porcine reproductive and respiratory syndrome virus 1 in China from 1994 to 2024.Virology. 2025 Apr;605:110457. doi: 10.1016/j.virol.2025.110457. Epub 2025 Feb 20. Virology. 2025. PMID: 39999587 Review.
Cited by
-
Development of SYBR green I-based real-time qPCR differential diagnosis assays for porcine reproductive and respiratory syndrome virus typing in Guangdong province.Front Vet Sci. 2025 Mar 5;12:1495128. doi: 10.3389/fvets.2025.1495128. eCollection 2025. Front Vet Sci. 2025. PMID: 40110430 Free PMC article.
-
The commercial PRRSV attenuated vaccine can be a potentially effective live trivalent vaccine vector.Appl Microbiol Biotechnol. 2025 May 2;109(1):109. doi: 10.1007/s00253-025-13502-5. Appl Microbiol Biotechnol. 2025. PMID: 40316839 Free PMC article.
-
Investigating the Presence and Genetic Variability of Porcine Circovirus Types 2 and 3 in Live Markets in Border Cities of Northeast China.Transbound Emerg Dis. 2025 Jun 19;2025:5526645. doi: 10.1155/tbed/5526645. eCollection 2025. Transbound Emerg Dis. 2025. PMID: 40574984 Free PMC article.
References
-
- Wang P., Zhang J., Ha Z., Xie C., Zhang H., Shi N., Han J., Xie Y., Li Z., Qiu X., et al. Synergistic Pathogenicity by Coinfection and Sequential Infection with JXA1-like HP-PRRSV and PCV2d in PCV2 Antibody-Positive Post-Weaned Pigs. Microb. Pathog. 2022;173:105810. doi: 10.1016/j.micpath.2022.105810. - DOI - PubMed
-
- Cui X., Xia D., Huang X., Sun Y., Shi M., Zhang J., Li G., Yang Y., Wang H., Cai X., et al. Analysis of Recombinant Characteristics Based on 949 PRRSV-2 Genomic Sequences Obtained from 1991 to 2021 Shows That Viral Multiplication Ability Contributes to Dominant Recombination. Microbiol. Spectr. 2022;10:e02934-22. doi: 10.1128/spectrum.02934-22. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

