Alteration of Gene Expression After Entecavir and Pegylated Interferon Therapy in HBV-Infected Chimeric Mouse Liver
- PMID: 39599858
- PMCID: PMC11598975
- DOI: 10.3390/v16111743
Alteration of Gene Expression After Entecavir and Pegylated Interferon Therapy in HBV-Infected Chimeric Mouse Liver
Abstract
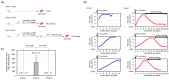
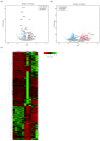
Cross-sectional analyses using liver tissue from chronic hepatitis B patients make it difficult to exclude the influence of host immune responses. In this study, we performed next-generation sequencing using the livers of hepatitis B virus (HBV)-infected uPA/SCID mice with humanized livers before and after antiviral therapy (AVT) with entecavir and pegylated interferon, and then performed a comparative transcriptome analysis of gene expression alteration. After HBV infection, the expression of genes involved in multiple pathways was significantly altered in the HBV-infected livers. After AVT, the levels of 37 out of 89 genes downregulated by HBV infection were restored, and 54 of 157 genes upregulated by HBV infection were suppressed. Interestingly, genes associated with hypoxia and KRAS signaling were included among the 54 genes upregulated by HBV infection and downregulated by AVT. Several genes associated with cell growth or carcinogenesis via hypoxia and KRAS signaling were significantly downregulated by AVT, with a potential application for the suppression of hepato-carcinogenesis.
Keywords: antiviral therapy; gene expression; hepatitis B virus; human hepatocyte; next-generation sequencing.
Conflict of interest statement
The authors declare no conflicts of interest. All the authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures



References
-
- World Health Organization . Global Progress Report on HIV, Viral Hepatitis and Sexually Transmitted Infections. World Health Organization; Geneva, Switzerland: 2021.
-
- Kumada H., Okanoue T., Onji M., Moriwaki H., Izumi N., Tanaka E., Chayama K., Sakisaka S., Takehara T., Oketani M., et al. Guidelines for the treatment of chronic hepatitis and cirrhosis due to hepatitis B virus infection for the fiscal year 2008 in Japan. Hepatol. Res. 2010;40:1–7. doi: 10.1111/j.1872-034X.2009.00633.x. - DOI - PubMed
-
- Lee Y., Suh D.J., Lim Y., Jung S.W., Kim K.M., Lee H.C., Chung Y., Lee Y.S., Yoo W., Kim S. Increased risk of adefovir resistance in patients with lamivudine-resistant chronic hepatitis B after 48 weeks of adefovir dipivoxil monotherapy. Hepatology. 2006;43:1385–1391. doi: 10.1002/hep.21189. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

