The Transcriptional Program of Staphylococcus aureus Phage K Is Affected by a Host rpoC Mutation That Confers Phage K Resistance
- PMID: 39599887
- PMCID: PMC11598898
- DOI: 10.3390/v16111773
The Transcriptional Program of Staphylococcus aureus Phage K Is Affected by a Host rpoC Mutation That Confers Phage K Resistance
Abstract
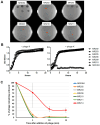
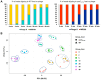
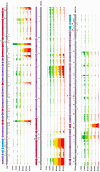
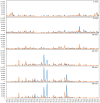
To better understand host-phage interactions and the genetic bases of phage resistance in a model system relevant to potential phage therapy, we isolated several spontaneous mutants of the USA300 S. aureus clinical isolate NRS384 that were resistant to phage K. Six of these had a single missense mutation in the host rpoC gene, which encodes the RNA polymerase β' subunit. To examine the hypothesis that mutations in the host RNA polymerase affect the transcription of phage genes, we performed RNA-seq analysis on total RNA samples collected from NRS384 wild-type (WT) and rpoCG17D mutant cultures infected with phage K, at different timepoints after infection. Infection of the WT host led to a steady increase of phage transcription relative to the host. Our analysis allowed us to define 53 transcriptional units and to categorize genes based on their temporal expression patterns. Predicted promoter sequences defined by conserved -35, -10, and, in some cases, extended -10 elements, were found upstream of early and middle genes. However, in many cases, sequences upstream of late genes did not contain clear, complete, canonical promoter sequences, suggesting that factors in addition to host RNA polymerase are required for their expression. Infection of the rpoCG17D mutant host led to a transcriptional pattern that was similar to that of the WT at early timepoints. However, beginning at 20 min after infection, transcription of late genes (such as phage structural genes and host lysis genes) was severely reduced. Our data indicate that the rpoCG17D mutation prevents the expression of phage late genes, resulting in a failed infection cycle for phage K. In addition to illuminating the global transcriptional landscape of phage K throughout the infection cycle, this study will inform our investigations into the basis of phage K's control of its transcriptional program as well as mechanisms of phage resistance.
Keywords: RNA-sequencing; bacteriophages; methicillin resistant Staphylococcus aureus; phage resistance; phage transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Global Transcriptomic Analysis of Bacteriophage-Host Interactions between a Kayvirus Therapeutic Phage and Staphylococcus aureus.Microbiol Spectr. 2022 Jun 29;10(3):e0012322. doi: 10.1128/spectrum.00123-22. Epub 2022 Apr 18. Microbiol Spectr. 2022. PMID: 35435752 Free PMC article.
-
Screening for Highly Transduced Genes in Staphylococcus aureus Revealed Both Lateral and Specialized Transduction.Microbiol Spectr. 2022 Feb 23;10(1):e0242321. doi: 10.1128/spectrum.02423-21. Epub 2022 Feb 9. Microbiol Spectr. 2022. PMID: 35138167 Free PMC article.
-
Genes Influencing Phage Host Range in Staphylococcus aureus on a Species-Wide Scale.mSphere. 2021 Jan 13;6(1):e01263-20. doi: 10.1128/mSphere.01263-20. mSphere. 2021. PMID: 33441407 Free PMC article.
-
Transcriptional Landscapes of Herelleviridae Bacteriophages and Staphylococcus aureus during Phage Infection: An Overview.Viruses. 2023 Jun 23;15(7):1427. doi: 10.3390/v15071427. Viruses. 2023. PMID: 37515114 Free PMC article. Review.
-
Genomics of staphylococcal Twort-like phages--potential therapeutics of the post-antibiotic era.Adv Virus Res. 2012;83:143-216. doi: 10.1016/B978-0-12-394438-2.00005-0. Adv Virus Res. 2012. PMID: 22748811 Review.
References
-
- Gorwitz R.J., Kruszon-Moran D., McAllister S.K., McQuillan G., McDougal L.K., Fosheim G.E., Jensen B.J., Killgore G., Tenover F.C., Kuehnert M.J. Changes in the Prevalence of Nasal Colonization with Staphylococcus aureus in the United States, 2001–2004. J. Infect. Dis. 2008;197:1226–1234. doi: 10.1086/533494. - DOI - PubMed
-
- Francis J.S., Doherty M.C., Lopatin U., Johnston C.P., Sinha G., Ross T., Cai M., Hansel N.N., Perl T., Ticehurst J.R., et al. Severe Community-Onset Pneumonia in Healthy Adults Caused by Methicillin-Resistant Staphylococcus aureus Carrying the Panton-Valentine Leukocidin Genes. Clin. Infect. Dis. 2005;40:100–107. doi: 10.1086/427148. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- AAI20020-001-00000/Interagency agreement between the United States Food and Drug Administration, National Institute of Allergy and Infectious Disease, Division of Microbiology and Infectious Diseases and National Institutes of Health
- Intramural Research Program/NIH - National Institute of Diabetes and Digestive and Kidney Diseases
- AAI20020-001/NIH NIAID
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

