STAT2/SLC27A3/PINK1-Mediated Mitophagy Remodeling Lipid Metabolism Contributes to Pazopanib Resistance in Clear Cell Renal Cell Carcinoma
- PMID: 39600540
- PMCID: PMC11588985
- DOI: 10.34133/research.0539
STAT2/SLC27A3/PINK1-Mediated Mitophagy Remodeling Lipid Metabolism Contributes to Pazopanib Resistance in Clear Cell Renal Cell Carcinoma
Abstract
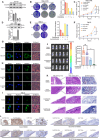
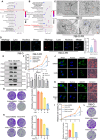
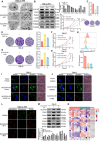
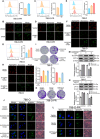
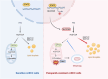
Background: Clear cell renal cell carcinoma (ccRCC) is a prevalent malignant tumor of the urinary system. While tyrosine kinase inhibitors (TKIs) are currently the first-line treatments for advanced/metastatic ccRCC, patients often develop resistance after TKI therapy. Lipid metabolic reprogramming, a hallmark of tumor progression, contributes to acquired drug resistance in various malignant tumors. Mitophagy, a process that maintains mitochondrial homeostasis, aids tumor cells in adapting to microenvironmental changes and consequently developing drug resistance. Solute carrier family 27 member 3 (SLC27A3), highly expressed in lipid-rich tumors like ccRCC, has been associated with poor prognosis. However, the impact of SLC27A3 and the transcription factor complex containing STAT2 on lipid metabolic reprogramming, mitophagy in ccRCC, and their role in TKI resistance remain unexplored. Methods: 786-O to pazopanib resistance was induced by gradient increase of concentration, and the genes related to lipid metabolism were screened by RNA sequencing. Bioinformatics was used to analyze the differential expression of SLC27A3 and its effect on patient prognosis, and to predict the activated pathway in pazopanib-resistant cells. Lipid droplets (LDs) were detected by Red Oil O and BODIPY probe. Micro-targeted lipidomic of acyl-coenzyme A (CoA) and lipid metabolomics were performed to screen potential metabolites of SLC27A3. The differential expression of SLC27A3 was detected in clinical samples. The differential expression of SLC27A3 and its effect on drug resistance of ccRCC tumor were detected in vitro and in vivo. Mitophagy was detected by electron microscopy, Mtphagy probe, and Western blot. The mitochondrial membrane potential (MMP) and reactive oxygen species (ROS) levels were detected by JC-1 and DCF probes. The binding site of the transcription factor complex to the SLC27A3 promoter was detected by dual-luciferase reporter gene assay. Results: SLC27A3, highly expressed in lipid-rich tumors such as ccRCC and glioblastoma, predicts poor prognosis. SLC27A3 expression level also increased in pazopanib-resistant 786-O cells (786-O-PR) with more LD accumulation compared to parental cells. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis from RNA sequencing showed that PINK1/Parkin-mediated mitophagy pathway was enriched in 786-O-PR. Knockdown of SLC27A3 markedly suppressed LD accumulation and mitophagy, and overcame pazopanib resistance in vitro and in vivo. Moreover, SLC27A3 functions as an acyl-CoA ligase catalyzing the formation of acyl-CoA, which refers to fatty acid oxidation accompanied by ROS production and synthesis of lipid. Overproduced acyl-CoA oxidation in mitochondria resulted in MMP decrease and amounts of ROS production, subsequently triggering PINK1/Parkin-mediated mitophagy. Moreover, mitophagy inhibition led to more ROS accumulation and cell death, indicating that mitophagy can keep ROS at an appropriate level by negative feedback. Mitophagy, simultaneously, prevented fatty acid oxidation in mitochondria by consuming CPT1A, forcing synthesis of triglycerides and cholesterol esters stored in LDs by transforming acyl-CoA, to support ccRCC progression. Besides, we found that STAT2 expression was positively correlated to SLC27A3. Transcriptional factor complex containing STAT2 could bind to the promoter of SLC27A3 mRNA to promote SLC27A3 transcription proved by dual-luciferase reporter assay, which also regulated LD metabolism and activated mitophagy during pazopanib resistance. Conclusion: SLC27A3 is up-regulated in pazopanib-resistant ccRCC and predicts poor prognosis. High expression of SLC27A3 produces excessive metabolites of various long-chain fatty acyl-CoA (12:0-, 16:0-, 17:0-, 20:3-CoA) to enter mitochondria for β-oxidation and produce amounts of ROS activating mitophagy. Subsequent mitophagy/ROS negative feedback controls ROS homeostasis and consumes CPT1A protein within mitochondria to suppress fatty acid β-oxidation, forcing acyl-CoA storage in LDs, mediating pazopanib resistance in ccRCC. Furthermore, STAT2 was identified as a core component of a potential upstream transcriptional factor complex for SLC27A3. Our findings shed new light on the underlying mechanism of SLC27A3 in ccRCC TKI resistance, which may provide a novel therapeutic target for the management of ccRCC.
Copyright © 2024 Dingheng Lu et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures


References
-
- Zhang X, Zhang G, Xu L, Bai X, Zhang J, Chen L, Lu X, Yu S, Jin Z, Sun H. Prediction of World Health Organization/International Society of Urological Pathology (WHO/ISUP) pathological grading of clear cell renal cell carcinoma by dual-layer spectral CT. Acad Radiol. 2023;30(10):2321–2328. - PubMed
-
- Huang B, Ren J, Ma Q, Yang F, Pan X, Zhang Y, Liu Y, Wang C, Zhang D, Wei L, et al. A novel peptide PDHK1-241aa encoded by circPDHK1 promotes ccRCC progression via interacting with PPP1CA to inhibit AKT dephosphorylation and activate the AKT-mTOR signaling pathway. Mol Cancer. 2024;23(1):34. - PMC - PubMed
-
- Rosellini M, Marchetti A, Mollica V, Rizzo A, Santoni M, Massari F. Prognostic and predictive biomarkers for immunotherapy in advanced renal cell carcinoma. Nat Rev Urol. 2023;20(3):133–157. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

