Targeting the NOTCH2/ADAM10/TCF7L2 Axis-Mediated Transcriptional Regulation of Wnt Pathway Suppresses Tumor Growth and Enhances Chemosensitivity in Colorectal Cancer
- PMID: 39601111
- PMCID: PMC11744699
- DOI: 10.1002/advs.202405758
Targeting the NOTCH2/ADAM10/TCF7L2 Axis-Mediated Transcriptional Regulation of Wnt Pathway Suppresses Tumor Growth and Enhances Chemosensitivity in Colorectal Cancer
Abstract
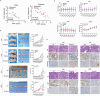
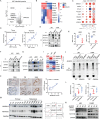
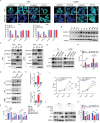
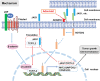
Wnt/β-catenin/transcription factor (TCF) transcriptional activity plays an integral role in colorectal cancer (CRC) carcinogenesis. However, to date, no drugs targeting this pathway are used in clinical practice owing to the undesirable and serious side effects. In this study, it is found that the transcriptional regulation of Wnt pathway is activated and associated with liver metastasis in CRC. Through high-throughput screening of 24 inhibitors on 12 CRC and three colorectal organoids in this organoid living biobank, adavivint is found to exhibit anti-tumor activity and low toxicity in colorectal organoids, independent of the canonical Wnt/β-catenin signaling. Mechanistically, ADAM10 is screened as a target of adavivint to specifically regulate the protein expression of NOTCH2, which mediates the transcriptional regulation of the Wnt pathway. NOTCH2 not directly interact with TCF7-like 2 (TCF7L2), a key downstream transcriptional factor of canonical Wnt/β-catenin signaling, but directly activated the transcription of TCF7L2 and Wnt target genes, such as MYC, JUN and CCND1/2. Furthermore, use of adavivint or blockage of ADAM10/NOTCH2/TCF7L2 signaling enhances the chemosensitivity of CRC cells. Overall, this study provides a promising candidate for the development of small-molecule inhibitors and reveals a potential therapeutic target for CRC.
Keywords: ADAM10; NOTCH2; TCF7L2; Wnt pathway; colorectal cancer; liver metastasis; organoid.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Siegel R. L., Wagle N. S., Cercek A., Smith R. A., Jemal A., CA Cancer J. Clin. 2023, 73, 233. - PubMed
-
- Cancer Genome Atlas Network , Nature 2012, 487, 330. - PubMed
-
- Tang J., Lam G. T., Brooks R D., Miles M., Useckaite Z., Johnson I Rd., Ung B S.‐Y., Martini C., Karageorgos L., Hickey S M., Selemidis S., Hopkins A M., Rowland A., Vather R., O'Leary J J., Brooks D A., Caruso M C., Logan J M., Cancer Lett. 2024, 585, 216639. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
