Decarboxylation of the Catalytic Lysine Residue by the C5α-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the OXA-58 Carbapenemase
- PMID: 39601221
- PMCID: PMC11972452
- DOI: 10.1021/acsinfecdis.4c00671
Decarboxylation of the Catalytic Lysine Residue by the C5α-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the OXA-58 Carbapenemase
Abstract
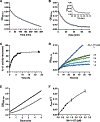
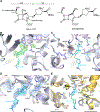
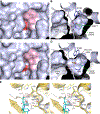
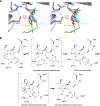
Antibiotic resistance in bacteria is a major global health concern. The wide spread of carbapenemases, bacterial enzymes that degrade the last-resort carbapenem antibiotics, is responsible for multidrug resistance in bacterial pathogens and has further significantly exacerbated this problem. Acinetobacter baumannii is one of the leading nosocomial pathogens due to the acquisition and wide dissemination of carbapenem-hydrolyzing class D β-lactamases, which have dramatically diminished available therapeutic options. Thus, new antibiotics that are active against multidrug-resistantA. baumannii and carbapenemase inhibitors are urgently needed. Here we report characterization of the interaction of the C5α-methyl-substituted carbapenem NA-1-157 with one of the clinically important class D carbapenemases, OXA-58. Antibiotic susceptibility testing shows that the compound is more potent than commercial carbapenems against OXA-58-producingA. baumannii, with a clinically sensitive MIC value of 1 μg/mL. Kinetic studies demonstrate that NA-1-157 is a very poor substrate of the enzyme due mainly to a significantly reduced deacylation rate. Mass spectrometry analysis shows that inhibition of OXA-58 by NA-1-157 proceeds through both the classical acyl-enzyme intermediate and a reversible covalent species. Time-resolved X-ray crystallographic studies reveal that upon acylation of the enzyme, the compound causes progressive decarboxylation of the catalytic lysine residue, thus severely impairing deacylation. Overall, this study demonstrates that the carbapenem NA-1-157 is highly resistant to degradation by the OXA-58 carbapenemase.
Keywords: Acinetobacter; OXA-58 carbapenemase; X-ray structure; antibiotic resistance; inhibitor.
Conflict of interest statement
The authors declare no competing financial interest.
The apo-OXA-58 and its complexes with NA-1–157 structure factors and atomic coordinates have been deposited in the PDB with PDB codes 9d78, 9d79, 9d7a, 9d7b, 9d7c, 9d7d and 9d8c. The authors will release the atomic coordinates and experimental data upon article publication.
Figures
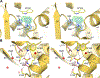
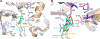
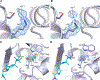
References
-
- Murray CJL; Ikuta KS; Sharara F; Swetschinski L; Robles Aguilar G; Gray A; Han C; Bisignano C; Rao P; Wool E; Johnson SC; Browne AJ; Chipeta MG; Fell F; Hackett S; Haines-Woodhouse G; Kashef Hamadani BH; Kumaran EAP; McManigal B; Achalapong S; Agarwal R; Akech S; Albertson S; Amuasi J; Andrews J; Aravkin A; Ashley E; Babin F-X; Bailey F; Baker S; Basnyat B; Bekker A; Bender R; Berkley JA; Bethou A; Bielicki J; Boonkasidecha S; Bukosia J; Carvalheiro C; Castañeda-Orjuela C; Chansamouth V; Chaurasia S; Chiurchiù S; Chowdhury F; Clotaire Donatien R; Cook AJ; Cooper B; Cressey TR; Criollo-Mora E; Cunningham M; Darboe S; Day NPJ; De Luca M; Dokova K; Dramowski A; Dunachie SJ; Duong Bich T; Eckmanns T; Eibach D; Emami A; Feasey N; Fisher-Pearson N; Forrest K; Garcia C; Garrett D; Gastmeier P; Giref AZ; Greer RC; Gupta V; Haller S; Haselbeck A; Hay SI; Holm M; Hopkins S; Hsia Y; Iregbu KC; Jacobs J; Jarovsky D; Javanmardi F; Jenney AWJ; Khorana M; Khusuwan S; Kissoon N; Kobeissi E; Kostyanev T; Krapp F; Krumkamp R; Kumar A; Kyu HH; Lim C; Lim K; Limmathurotsakul D; Loftus MJ; Lunn M; Ma J; Manoharan A; Marks F; May J; Mayxay M; Mturi N; Munera-Huertas T; Musicha P; Musila LA; Mussi-Pinhata MM; Naidu RN; Nakamura T; Nanavati R; Nangia S; Newton P; Ngoun C; Novotney A; Nwakanma D; Obiero CW; Ochoa TJ; Olivas-Martinez A; Olliaro P; Ooko E; Ortiz-Brizuela E; Ounchanum P; Pak GD; Paredes JL; Peleg AY; Perrone C; Phe T; Phommasone K; Plakkal N; Ponce-de-Leon A; Raad M; Ramdin T; Rattanavong S; Riddell A; Roberts T; Robotham JV; Roca A; Rosenthal VD; Rudd KE; Russell N; Sader HS; Saengchan W; Schnall J; Scott JAG; Seekaew S; Sharland M; Shivamallappa M; Sifuentes-Osornio J; Simpson AJ; Steenkeste N; Stewardson AJ; Stoeva T; Tasak N; Thaiprakong A; Thwaites G; Tigoi C; Turner C; Turner P; van Doorn HR; Velaphi S; Vongpradith A; Vongsouvath M; Vu H; Walsh T; Walson JL; Waner S; Wangrangsimakul T; Wannapinij P; Wozniak T; Young Sharma TEMW; Yu KC; Zheng P; Sartorius B; Lopez AD; Stergachis A; Moore C; Dolecek C; Naghavi M Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022, 399, 629–655. - PMC - PubMed
-
- O’Neill J Tackling drug-resistant infections globally: final report and recommendations. The Review on Antimicrobial Resistance; Government of the United Kingdom, 2016. (accessed on July 12, 2024).
-
- Rice LB Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

