LINE-1 hypomethylation characterizes the inflammatory response in coeliac disease associated-intestinal mucosa and small bowel adenocarcinomas
- PMID: 39601242
- PMCID: PMC11638666
- DOI: 10.1002/path.6371
LINE-1 hypomethylation characterizes the inflammatory response in coeliac disease associated-intestinal mucosa and small bowel adenocarcinomas
Abstract
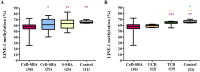
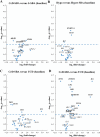
Long interspersed nuclear elements 1 (LINE-1) are the most abundant and the only autonomous mobile elements in the human genome. When their epigenetic repression is removed, it can lead to disease, such as autoimmune diseases and cancer. Coeliac disease (CeD) is an immune-mediated disease triggered by an abnormal T-cell response to dietary gluten and a predisposing condition of small bowel adenocarcinoma (SBA), frequently characterized by epigenetic alterations. The aim of this work was to assess LINE-1 methylation by bisulphite pyrosequencing and NanoString® gene transcription analysis in 38 CeD-SBAs compared with 25 SBAs associated with Crohn's disease (CrD-SBAs) and 25 sporadic SBAs (S-SBA). Both analyses were also performed in duodenal mucosae from 12 untreated CeD patients (UCD) and 19 treated CeD patients (TCD), and in 11 samples of normal intestinal mucosa to better investigate the role of LINE-1 deregulation in CeD and in CeD-SBA. A significant loss of LINE-1 methylation was observed in CeD-SBAs and in mucosae from UCD patients (with very similar methylation levels) compared with controls. By contrast, a restoration of normal LINE-1 methylation levels was found in TCD mucosae after a strict gluten-free diet. LINE-1 hypomethylation does not lead to expression of ORF1 and ORF2, with the only exception being for one CeD-SBA. The expression analysis of enzymes modulating DNA methylation and inflammatory genes confirmed that CeD-SBA shared a very similar expression profile of UCD mucosae showing a strong upregulation of genes involved in inflammation, immune response, and T-cell activity compared with TCD mucosae. For the first time, this work demonstrates that loss of DNA methylation is an intrinsic epigenetic feature of CeD, accompanying the immune response as a reversible mechanism in patients following a strict gluten-free diet, and suggests the possible role of LINE-1 hypomethylation in promoting cell adaptability during the gliadin-related inflammatory process. © 2024 The Author(s). The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: LINE‐1 hypomethylation; LINE‐1 retrotransposition; coeliac disease; immune‐mediated disorders; small bowel adenocarcinomas.
© 2024 The Author(s). The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Figures

References
-
- Streicher F, Jouvenet N. Stimulation of innate immunity by host and viral RNAs. Trends Immunol 2019; 40: 1134–1148. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

