The respective roles of TMPRSS2 and cathepsins for SARS-CoV-2 infection in human respiratory organoids
- PMID: 39601592
- PMCID: PMC11784140
- DOI: 10.1128/jvi.01853-24
The respective roles of TMPRSS2 and cathepsins for SARS-CoV-2 infection in human respiratory organoids
Abstract
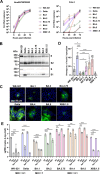
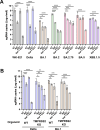
A critical aspect of the mechanism of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is the protease-mediated activation of the viral spike (S) protein. The type II transmembrane serine protease TMPRSS2 is crucial for SARS-CoV-2 infection in lung epithelial Calu-3 cells and murine airways. However, the importance of TMPRSS2 needs to be re-examined because the ability to utilize TMPRSS2 is significantly reduced in the Omicron variants that spread globally. For this purpose, replication profiles of SARS-CoV-2 were analyzed in human respiratory organoids. All tested viruses, including Omicron variants, replicated efficiently in these organoids. Notably, all SARS-CoV-2 strains retained replication ability in TMPRSS2-gene knockout (KO) respiratory organoids, suggesting that TMPRSS2 is not essential for SARS-CoV-2 infection in human respiratory tissues. However, TMPRSS2-gene knockout significantly reduces the inhibitory effect of nafamostat, indicating the advantage of TMPRSS2-utilizing ability for the SARS-CoV-2 infection in these organoids. Interestingly, Omicron variants regained the TMPRSS2-utilizing ability in recent subvariants. The basal infectivity would be supported mainly by cathepsins because the cathepsin inhibitor, EST, showed a significant inhibitory effect on infection with any SARS-CoV-2 strains, mainly when used with nafamostat. A supplementary contribution of other serine proteases was also suggested because the infection of the Delta variant was still inhibited partially by nafamostat in TMPRSS2 KO organoids. Thus, various proteases, including TMPRSS2, other serine proteases, and cathepsins, co-operatively contribute to SARS-CoV-2 infection significantly in the respiratory organoids. Thus, SARS-CoV-2 infection in the human respiratory tissues would be more complex than observed in cell lines or mice.
Importance: We explored how the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus infects human respiratory organoids, which are a cultured cell model made to mimic the physiological conditions of the human airways. We focused on understanding the role of different proteases of host cells in activating the virus spike proteins. Specifically, we looked at TMPRSS2, a transmembrane serine protease, and cathepsin L, a lysosomal enzyme, which helps the virus enter cells by cutting the viral spike protein. We discovered that while TMPRSS2 is crucial for the virus in certain cells and animal models, other proteases, including cathepsins and various serine proteases, also play significant roles in the SARS-CoV-2 infection of human respiratory organoids. We suggest that SARS-CoV-2 uses a more complex mechanism involving multiple proteases to infect human airways, differing from what we see in conventional cell lines or animal models. This complexity might help explain how different variants can spread and infect people effectively.
Keywords: SARS-CoV-2; TMPRSS2; cathepsin; cleavage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- JP243fa627001/Japan Agency for Medical Research and Development (AMED)
- Astellas Foundation for Research on Metabolic Disorders
- JP21gm1610005/Japan Agency for Medical Research and Development (AMED)
- JP23jf0126002/Japan Agency for Medical Research and Development (AMED)
- 23K17416/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

