Transcriptomic and genetic analysis reveals a Zn2Cys6 transcription factor specifically required for conidiation in submerged cultures of Thermothelomyces thermophilus
- PMID: 39601596
- PMCID: PMC11708020
- DOI: 10.1128/mbio.03111-24
Transcriptomic and genetic analysis reveals a Zn2Cys6 transcription factor specifically required for conidiation in submerged cultures of Thermothelomyces thermophilus
Abstract
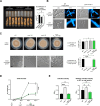
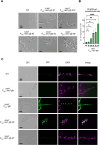
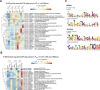
Filamentous fungi are important producers of enzymes and secondary metabolites. The industrial thermophilic species, Thermothelomyces thermophilus, is closely related to the model fungus, Neurospora crassa. A critical aspect of the filamentous fungal life cycle is the production of asexual spores (conidia), which are regulated by various stimuli, including nutrient availability. Several species of fungi, including T. thermophilus, produce conidia under submerged fermentation conditions, which can be detrimental to product yields. In this study, transcriptional profiling of T. thermophilus was used to map changes during asexual development in submerged cultures, which revealed commonalities of regulation between T. thermophilus and N. crassa. We further identified a transcription factor, res1, whose deletion resulted in a complete loss of conidia production under fermentation conditions, but which did not affect conidiation on plates. Under fermentation conditions, the ∆res1 deletion strain showed increased biomass production relative to the wild-type strain, indicating that the manipulation of res1 in T. thermophilus has the potential to increase productivity in industrial settings. Overexpression of res1 caused a severe growth defect and early conidia production on both plates and in submerged cultures, indicating res1 overexpression can bypass regulatory aspects associated with conidiation on plates. Using chromatin-immunoprecipitation sequencing, we identified 35 target genes of Res1, including known conidiation regulators identified in N. crassa, revealing common and divergent aspects of asexual reproduction in these two species.IMPORTANCEFilamentous fungi, such as Thermothelomyces thermophilus, are important industrial species and have been harnessed in the Biotechnology industry for the production of industrially relevant chemicals and proteins. However, under fermentation conditions, some filamentous fungi will undergo a switch from mycelial growth to asexual development. In this study, we use transcriptional profiling of asexual development in T. thermophilus and identify a transcription factor that specifically regulates the developmental switch to the production of unwanted asexual propagules under fermentation conditions, thus altering secreted protein production. Mutations in this transcription factor Res1 result in the loss of asexual development in submerged cultures but do not affect asexual sporulation when exposed to air. The identification of stage-specific developmental regulation of asexual spore production and comparative analyses of conidiation in filamentous ascomycete species have the potential to further manipulate these species for industrial advantage.
Keywords: Thermothelomyces; asexual development; conidiation; fungal biotechnology; transcriptional regulation.
Conflict of interest statement
The authors declare that the inhibition of conidia production via a ∆
Figures




References
-
- Liu Q, Gao R, Li J, Lin L, Zhao J, Sun W, Tian C. 2017. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol Biofuels 10:1. doi: 10.1186/s13068-016-0693-9 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

