The Hox protein Antennapedia orchestrates Drosophila adult flight muscle development
- PMID: 39602537
- PMCID: PMC11601212
- DOI: 10.1126/sciadv.adr2261
The Hox protein Antennapedia orchestrates Drosophila adult flight muscle development
Abstract
Muscle development and diversity require a large number of spatially and temporally regulated events controlled by transcription factors (TFs). Drosophila has long stood as a model to study myogenesis due to the highly conserved key TFs involved at all stages of muscle development. While many studies focused on the diversification of Drosophila larval musculature, how distinct adult muscle types are generated is much less characterized. Here, we identify an essential regulator of Drosophila thoracic flight muscle development, the Hox TF Antennapedia (Antp). Correcting a long-standing belief that flight muscle development occurs without the input of Hox TFs, we show that Antp intervenes at several stages of flight muscle development, from the establishment of the progenitor pool in the embryo to myoblast differentiation in the early pupa. Furthermore, the precisely regulated clearance of Hox in the developing flight muscle fibers is required to allow for fibrillar muscle fate diversification, setting these muscles apart from all other adult tubular muscle types.
Figures
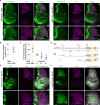

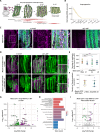

References
-
- Michelson A. M., Muscle pattern diversification in Drosophila is determined by the autonomous function of homeotic genes in the embryonic mesoderm. Development 120, 755–768 (1994). - PubMed
-
- Lawrence P. A., Johnston P., The genetic specification of pattern in a drosophila muscle. Cell 36, 775–782 (1984). - PubMed
-
- Capovilla M., Kambris Z., Botas J., Direct regulation of the muscle-identity gene apterous by a Hox protein in the somatic mesoderm. Development 128, 1221–1230 (2001). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

