Macrophages are activated toward phagocytic lymphoma cell clearance by pentose phosphate pathway inhibition
- PMID: 39603243
- PMCID: PMC11722127
- DOI: 10.1016/j.xcrm.2024.101830
Macrophages are activated toward phagocytic lymphoma cell clearance by pentose phosphate pathway inhibition
Abstract
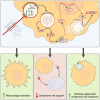
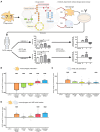
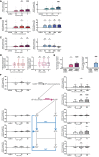
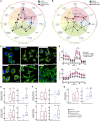
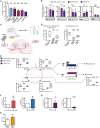
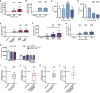
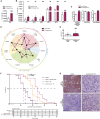
Macrophages in the B cell lymphoma microenvironment represent a functional node in progression and therapeutic response. We assessed metabolic regulation of macrophages in the context of therapeutic antibody-mediated phagocytosis. Pentose phosphate pathway (PPP) inhibition induces increased phagocytic lymphoma cell clearance by macrophages in vitro, in primary human chronic lymphocytic leukemia (CLL) patient co-cultures, and in mouse models. Addition of the PPP inhibitor S3 to antibody therapy achieves significantly prolonged overall survival in an aggressive B cell lymphoma mouse model. PPP inhibition induces metabolic activation and pro-inflammatory polarization of macrophages while it decreases macrophages' support for survival of lymphoma cells empowering anti-lymphoma function. As a mechanism of macrophage repolarization, the link between PPP and immune regulation was identified. PPP inhibition causes decreased glycogen level and subsequent modulation of the immune modulatory uridine diphosphate glucose (UDPG)-Stat1-Irg1-itaconate axis. Thus, we hypothesize the PPP as a key regulator and targetable modulator of macrophage activity in lymphoma to improve efficacy of immunotherapies and prolong survival.
Keywords: ADCP; Irg1; immunotherapy; itaconate; lymphoma; macrophage; metabolic modulation; pentose phosphate pathway; phagocytosis; polarization.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Cassetta L., Pollard J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018;17:887–904. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

