LungVis 1.0: an automatic AI-powered 3D imaging ecosystem unveils spatial profiling of nanoparticle delivery and acinar migration of lung macrophages
- PMID: 39604430
- PMCID: PMC11603200
- DOI: 10.1038/s41467-024-54267-1
LungVis 1.0: an automatic AI-powered 3D imaging ecosystem unveils spatial profiling of nanoparticle delivery and acinar migration of lung macrophages
Abstract
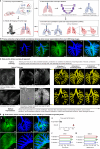
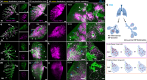
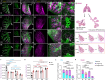
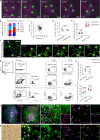
Targeted (nano-)drug delivery is essential for treating respiratory diseases, which are often confined to distinct lung regions. However, spatio-temporal profiling of drugs or nanoparticles (NPs) and their interactions with lung macrophages remains unresolved. Here, we present LungVis 1.0, an AI-powered imaging ecosystem that integrates light sheet fluorescence microscopy with deep learning-based image analysis pipelines to map NP deposition and dosage holistically and quantitatively across bronchial and alveolar (acinar) regions in murine lungs for widely-used bulk-liquid and aerosol-based delivery methods. We demonstrate that bulk-liquid delivery results in patchy NP distribution with elevated bronchial doses, whereas aerosols achieve uniform deposition reaching distal alveoli. Furthermore, we reveal that lung tissue-resident macrophages (TRMs) are dynamic, actively patrolling and redistributing NPs within alveoli, contesting the conventional paradigm of TRMs as static entities. LungVis 1.0 provides an advanced framework for exploring pulmonary delivery dynamics and deepening insights into TRM-mediated lung immunity.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- 686098/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 953183/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources
Miscellaneous

