Identification and characterization of the Quinoa AP2/ERF gene family and their expression patterns in response to salt stress
- PMID: 39604476
- PMCID: PMC11603269
- DOI: 10.1038/s41598-024-81046-1
Identification and characterization of the Quinoa AP2/ERF gene family and their expression patterns in response to salt stress
Abstract
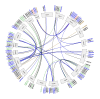
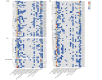
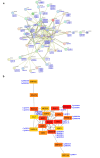
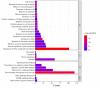
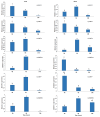
The APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) transcription factors play crucial roles in plant growth, development, and responses to biotic and abiotic stresses. This study was performed to comprehensively identify and characterize the AP2/ERF gene family in quinoa (Chenopodium quinoa Willd.), a highly resilient pseudocereal crop known for its salinity tolerance. A total of 150 CqAP2/ERF genes were identified in the quinoa genome; these genes were unevenly distributed across the chromosomes. Phylogenetic analysis divided the CqAP2/ERFs into five subfamilies: 71 ERF, 49 DREB, 23 AP2, 3 RAV, and 4 Soloist. Additionally, the DREB and ERF subfamilies were subdivided into four and seven subgroups, respectively. The exon-intron structure of the putative CqAP2/ERF genes and the conserved motifs of their encoded proteins were also identified, showing general conservation within the phylogenetic subgroups. Promoter analysis revealed many cis-regulatory elements associated with light, hormones, and response mechanisms within the promoter regions of CqAP2/ERF genes. Synteny analysis revealed that segmental duplication under purifying selection pressure was the major evolutionary driver behind the expansion of the CqAP2/ERF gene family. The protein-protein interaction network predicted the pivotal CqAP2/ERF proteins and their interactions involved in the regulation of various biological processes including stress response mechanisms. The expression profiles obtained from RNA-seq and qRT-PCR data detected several salt-responsive CqAP2/ERF genes, particularly from the ERF, DREB, and RAV subfamilies, with varying up- and downregulation patterns, indicating their potential roles in salt stress responses in quinoa. Overall, this study provides insights into the structural and evolutionary features of the AP2/ERF gene family in quinoa, offering candidate genes for further analysis of their roles in salt tolerance and molecular breeding.
Keywords: Chenopodium quinoa; AP2/ERF transcription factors; DREB; Gene Duplication; Regulatory Network; Salinity.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures





References
-
- Devkota, K. P., Devkota, M., Rezaei, M. & Oosterbaan, R. Managing salinity for sustainable agricultural production in salt-affected soils of irrigated drylands. Agric. Syst.198, 103390 (2022).
-
- Koca, Y. O. Effects of different salt concentrations on Quinoa Seedling Quality. Int. J. Second Metab.4, 20–26 (2017).
-
- Manaa, A. et al. Salinity tolerance of quinoa (Chenopodium quinoa Willd) as assessed by chloroplast ultrastructure and photosynthetic performance. Environ. Exp. Bot.162, 103–114 (2019).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

