Elevated expression of the retrotransposon LINE-1 drives Alzheimer's disease-associated microglial dysfunction
- PMID: 39604588
- PMCID: PMC11602836
- DOI: 10.1007/s00401-024-02835-6
Elevated expression of the retrotransposon LINE-1 drives Alzheimer's disease-associated microglial dysfunction
Abstract
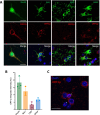
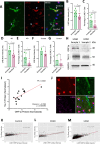
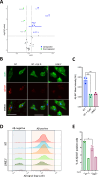
Aberrant activity of the retrotransposable element long interspersed nuclear element-1 (LINE-1) has been hypothesized to contribute to cellular dysfunction in age-related disorders, including late-onset Alzheimer's disease (LOAD). However, whether LINE-1 is differentially expressed in cell types of the LOAD brain, and whether these changes contribute to disease pathology is largely unknown. Here, we examined patterns of LINE-1 expression across neurons, astrocytes, oligodendrocytes, and microglia in human postmortem prefrontal cortex tissue from LOAD patients and cognitively normal, age-matched controls. We report elevated immunoreactivity of the open reading frame 1 protein (ORF1p) encoded by LINE-1 in microglia from LOAD patients and find that this immunoreactivity correlates positively with disease-associated microglial morphology. In human iPSC-derived microglia (iMG), we found that CRISPR-mediated transcriptional activation of LINE-1 drives changes in microglial morphology and cytokine secretion and impairs the phagocytosis of amyloid beta (Aβ). We also find LINE-1 upregulation in iMG induces transcriptomic changes genes associated with antigen presentation and lipid metabolism as well as impacting the expression of many AD-relevant genes. Our data posit that heightened LINE-1 expression may trigger microglial dysregulation in LOAD and that these changes may contribute to disease pathogenesis, suggesting a central role for LINE-1 activity in human LOAD.
Keywords: Alzheimer’s disease; LINE-1; Microglia; Neuroinflammation; Retrotransposons; Transposable elements.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare no competing interests. Ethics approval and consent to participate: The research was conducted in accordance with the guidelines of Institutional Review Board (IRB) of Columbia University New York under protocol AAAR4962. This study utilized induced pluripotent stem cell (iPSC)-derived microglia, hence no direct human participants were involved. Human iPSC lines C1-iPSC (Gibco, catalog A18945), C2-iPSC (ATCC, catalog no. ACS-1024), and CU-iPSC (Columbia Stem Cell Initiative core facility), were obtained in accordance with ethical guidelines for research. Consent for publication: This manuscript does not contain data from individuals necessitating consent for publication.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

