Proximal tubule cell maturation rate and function are controlled by PPARα signaling in kidney organoids
- PMID: 39604738
- PMCID: PMC11603349
- DOI: 10.1038/s42003-024-07069-6
Proximal tubule cell maturation rate and function are controlled by PPARα signaling in kidney organoids
Abstract
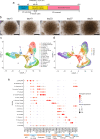
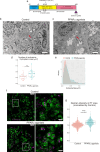
Human pluripotent stem cell-derived kidney organoids are expected to be a useful tool for new drug discoveries, however, the immaturation of kidney organoids causes difficulties in recapitulating renal pharmacokinetics using organoids. Here, we performed time-course single-cell RNA sequencing of kidney organoids and revealed cell heterogeneity in the maturation rate of the proximal tubule. An unbiased analysis to identify upstream targets of genes that are expressed differentially between cells with low and high maturation rates revealed a higher activation of PPARα signaling in rapidly maturing cells. Treatment with a combination of a PPARα agonist and an RXRα agonist induced genes related to proximal tubule maturation and increased the capacity for protein uptake as well as the sensitivity to nephrotoxicity by cisplatin. This method to promote the maturation rate of proximal tubule cells has the potential to be utilized in microphysiological systems to recapitulate proximal tubule functions and to screen nephrotoxic drugs.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature526, 564–568 (2015). - PubMed
-
- Shimizu, T. et al. A novel ADPKD model using kidney organoids derived from disease-specific human iPSCs. Biochem. Biophys. Res. Commun.529, 1186–1194 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

