MYB transcription factors in Peucedanum Praeruptorum Dunn: the diverse roles of the R2R3-MYB subfamily in mediating coumarin biosynthesis
- PMID: 39604839
- PMCID: PMC11604020
- DOI: 10.1186/s12870-024-05864-1
MYB transcription factors in Peucedanum Praeruptorum Dunn: the diverse roles of the R2R3-MYB subfamily in mediating coumarin biosynthesis
Abstract
Background: The MYB superfamily (v-myb avian myeloblastosis viral oncogene homolog) plays a role in plant growth and development, environmental stress defense, and synthesis of secondary metabolites. Little is known about the regulatory function of MYB genes in Peucedanum praeruptorum Dunn, although many MYB family members, especially R2R3-MYB genes, have been extensively studied in model plants.
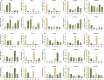
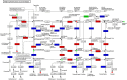
Results: A total of 157 R2R3-MYB transcription factors from P. praeruptorum were identified using bioinformatics analysis. Comprehensive analyses including chromosome location, microsynteny, gene structure, conserved motif, phylogenetic tree, and conserved domain were further performed. The length of the 157 transcription factors ranged from 120 to 1,688 amino acids (molecular weight between 14.21 and 182.69 kDa). All proteins were hydrophilic. Subcellular localization predictions showed that 155 PpMYB proteins were localized in the nucleus, with PpMYB12 and PpMYB157 localized in the chloroplasts and mitochondria, respectively. Ten conserved motifs were identified in the PpMYBs, all of which contained typical MYB domains. Transcriptome analysis identified 47,902 unigenes. Kyoto Encyclopedia of Genes and Genomes analysis revealed 136 pathways, of which 524 genes were associated with the phenylpropanoid pathway. Differential expressed genes (DEGs) before and after bolting showed that 11 genes were enriched in the phenylpropanoid pathway. Moreover, the expression patterns of transcription genes were further verified by qRT-PCR. With high-performance liquid chromatography (HPLC), 8 coumarins were quantified from the root, stem, and leaf tissue samples of P. praeruptorum at different stages. Praeruptorin A was found in both roots and leaves before bolting, whereas praeruptorin B was mainly concentrated in the roots, and the content of both decreased in the roots and stems after bolting. Praeruptorin E content was highest in the leaves and increased with plant growth. The correlation analysis between transcription factors and coumarin content showed that the expression patterns of PpMYB3 and PpMYB103 in roots align with the accumulation trends of praeruptorin A, praeruptorin B, praeruptorin E, scopoletin, and isoscopoletin, which declined in content after bolting, suggesting that these genes may positively regulate the biosynthesis of coumarins. Eleven distinct metabolites and 48 DEGs were identified. Correlation analysis revealed that the expression of all DEGs were significantly related to the accumulation of coumarin metabolites, indicating that these genes are involved in the regulation of coumarin biosynthesis.
Conclusions: R2R3-MYB transcription factors may be involved in the synthesis of coumarin. Our findings provide basic data and a rationale for future an in-depth studies on the role of R2R3-MYB transcription factors in the growth and regulation of coumarin synthesis.
Keywords: Peucedanum Praeruptorum Dunn; Coumarin; Phenylpropanoid pathway; R2R3-MYB; Transcriptional regulation.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The samples were collected from the Botanical Garden growing in West Anhui University. The voucher specimens were deposited in the herbarium of West Anhui University and identified by Prof. Bangxing Han. This study complies with relevant institutional, national, and international guidelines and legislation. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures









References
-
- Song C, Li X, Jia B, Liu L, Ou J, Han B. De novo Transcriptome sequencing coupled with co-expression analysis reveal the transcriptional regulation of key genes involved in the formation of active ingredients in Peucedanum Praeruptorum Dunn under bolting period. Front Genet. 2021;12:683037. - DOI - PMC - PubMed
-
- Song Y, Jing W, Yan R, Wang Y. Research progress of the studies on the roots of Peucedanum Praeruptorum dunn (Peucedani radix). Pak J Pharm Sci. 2015;28(1):71–81. - PubMed
-
- Song Y, Jing W, Yang F, Shi Z, Yao M, Yan R, Wang Y. Simultaneously enantiospecific determination of (+)-trans-khellactone, (+/-)-praeruptorin A, (+/-)-praeruptorin B, (+)-praeruptorin E, and their metabolites, (+/-)-cis-khellactone, in rat plasma using online solid phase extraction-chiral LC-MS/MS. J Pharm Biomed Anal. 2014;88:269–77. - DOI - PubMed
-
- Xie J, Tang X, Xie C, Wang Y, Huang J, Jin J, Liu H, Zhong C, Zhou R, Ren G, et al. Comparative analysis of root anatomical structure, chemical components and differentially expressed genes between early bolting and unbolting in Peucedanum Praeruptorum Dunn. Genomics. 2023;115(2):110557. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

