This is a preprint.
Single-cell spatial transcriptomics of fixed, paraffin-embedded biopsies reveals colitis-associated cell networks
- PMID: 39605355
- PMCID: PMC11601222
- DOI: 10.1101/2024.11.11.623014
Single-cell spatial transcriptomics of fixed, paraffin-embedded biopsies reveals colitis-associated cell networks
Abstract
Background & aims: Imaging-based, single-cell spatial transcriptomics (iSCST) using formalin-fixed, paraffin-embedded (FFPE) tissue could transform translational research by retaining all tissue cell subsets and spatial locations while enabling the analysis of archived specimens. We aimed to develop a robust framework for applying iSCST to archived clinical FFPE mucosal biopsies from patients with inflammatory bowel disease (IBD).
Methods: We performed a comprehensive benchmarking comparison of three iSCST platforms capable of analyzing FFPE specimens. We analyzed FFPE mucosal biopsies (n=57) up to 5 years old from non-IBD controls (HC; n=9) and patients with ulcerative colitis (UC;n=11). After platform-specific cell segmentation, we applied a uniform data processing pipeline to all datasets, including transcript detection, cell annotation, differential gene expression, and neighborhood enrichment. Transcriptomic signatures identified with iSCST were validated using external, publicly available bulk transcriptomic datasets.
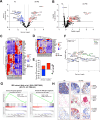
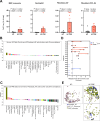
Results: A custom 290-plex Xenium gene panel exhibited the highest sensitivity and specificity for transcript detection, enabling precise identification and quantification of diverse cell subsets and differentially expressed genes across cell types and disease states. We mapped transcriptionally distinct fibroblast subsets to discrete spatial locations and identified inflammation-associated fibroblasts (IAFs) and monocytes as a colitis-associated cellular neighborhood. We also identified signatures associated with Vedolizumab (VDZ) responsiveness. VDZ non-responders were characterized by an IAF-monocyte transcriptional signature, while responders exhibited enrichment of epithelial gene sets.
Conclusions: Our optimized iSCST framework for archived FFPE biopsies provides unique advantages for assessing the role of colitis-associated cellular networks in routinely collected clinical samples. FFPE-based biomarkers could integrate with existing clinical workflows and potentially aid in risk-stratifying patients.
Keywords: Spatial transcriptomics; ulcerative colitis; vedolizumab.
Conflict of interest statement
Competing interests statement The Kattah, Combes and Fragiadakis lab receive research support from Eli Lilly for work unrelated to this manuscript. The Combes lab receive research support from Genentech for work unrelated to this manuscript. MGK is a member of the scientific advisory boards of Santa Ana Bio and Switchback Therapeutics and has received consulting fees from Cellarity, Spyre Therapeutics, Morphic Therapeutic, Sonoma Biotherapeutics, and Surrozen. AJC is a member of the scientific advisory board of Foundery innovations and has received consulting fees from Survey Genomics. UM is a consultant for Abbvie, Janssen, Takeda, Pfizer, BMS, Gilead, Enveda, Lilly, Merck, Rani Therapeutics, Celltrion, Abivax and received grant support from Leona and Harry Helmsley Charitable Trust. DYO has received research support from Merck, PACT Pharma, the Parker Institute for Cancer Immunotherapy, Poseida Therapeutics, TCR2 Therapeutics, Roche/Genentech, and Nutcracker Therapeutics; travel and accommodations from Roche/Genentech and Poseida Therapeutics; and has consulted for Revelation Partners.
Figures



References
-
- Parikh K. et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567, 49–55 (2019). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
