This is a preprint.
SARS-CoV-2 ORF3a blocks lysosomal cholesterol egress by disrupting VPS39-regulated NPC2 trafficking and BMP metabolism
- PMID: 39605369
- PMCID: PMC11601339
- DOI: 10.1101/2024.11.13.623299
SARS-CoV-2 ORF3a blocks lysosomal cholesterol egress by disrupting VPS39-regulated NPC2 trafficking and BMP metabolism
Abstract
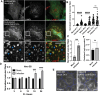
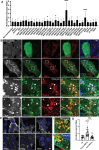
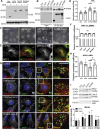
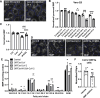
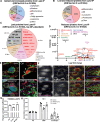
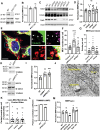
Cholesterol homeostasis relies on lysosomes, which release free cholesterol from degraded lipids. We show that SARS-CoV-2 blocks lysosomal cholesterol export through its protein ORF3a. ORF3a binds the HOPS subunit VPS39, and disrupting this interaction restores cholesterol trafficking. Two mechanisms underlie this defect. First, ORF3a-VPS39 interaction traps the sorting receptor CI-MPR and the retrieval complex retromer in endosomes/lysosomes, impairing trafficking of the cholesterol transporter NPC2. Retromer deletion reproduced these defects. Second, ORF3a reduces bis(monoacylglycerol)phosphates (BMPs), lysosomal lipids required for cholesterol export. Lipidomics and proteomics revealed altered metabolism of BMP precursors, mitochondrial phosphatidylglycerols (PGs), and reduced mitochondrial proteins at lysosomes. ORF3a-VPS39 interaction decreased lysosome-mitochondrion membrane contact sites (MCS), excluding autophagy or mitochondrion-derived vesicles as routes for PG transfer. VPS39 deletion decreased the MCS and BMPs. These findings identify VPS39 as a regulator of NPC2 trafficking and BMP biosynthesis and reveal that ORF3a disrupts both pathways to block cholesterol egress.
Keywords: Cellular cholesterol transport; HOPS; bis(monoacylglycerol)phosphates (BMPs); lysosome-mitochondrion interactions; retromer.
Figures


References
-
- Li Y., Zhang Y., Lu R., Dai M., Shen M., Zhang J., Cui Y., Liu B., Lin F., Chen L., Han D., Fan Y., Zeng Y., Li W., Li S., Chen X., Li H. & Pan P., Lipid metabolism changes in patients with severe COVID-19. Clin Chim Acta 2021. 517 33639119
-
- Rezaei A., Neshat S. & Heshmat-Ghahdarijani K., Alterations of Lipid Profile in COVID-19: A Narrative Review. Curr Probl Cardiol 2022. 47(3) 34272088
-
- Zickler M., Stanelle-Bertram S., Ehret S., Heinrich F., Lange P., Schaumburg B., Kouassi N. M., Beck S., Jaeckstein M. Y., Mann O., Krasemann S., Schroeder M., Jarczak D., Nierhaus A., Kluge S., Peschka M., Schlüter H., Renné T., Pueschel K., Kloetgen A., Scheja L., Ondruschka B., Heeren J. & Gabriel G., Replication of SARS-CoV-2 in adipose tissue determines organ and systemic lipid metabolism in hamsters and humans. Cell Metab 2022. 34(1) 34895500
-
- Xu E., Xie Y. & Al-Aly Z., Risks and burdens of incident dyslipidaemia in long COVID: a cohort study. Lancet Diabetes Endocrinol 2023. 11(2) 36623520
-
- López-Hernández Y., Oropeza-Valdez J. J., García Lopez D. A., Borrego J. C., Murgu M., Valdez J., López J. A. & Monárrez-Espino J., Untargeted analysis in post-COVID-19 patients reveals dysregulated lipid pathways two years after recovery. Front Mol Biosci 2023. 10 36936993
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
