This is a preprint.
Arginine Regulates the Mucoid Phenotype of Hypervirulent Klebsiella pneumoniae
- PMID: 39605402
- PMCID: PMC11601523
- DOI: 10.1101/2024.11.20.624485
Arginine Regulates the Mucoid Phenotype of Hypervirulent Klebsiella pneumoniae
Update in
-
Arginine regulates the mucoid phenotype of hypervirulent Klebsiella pneumoniae.Nat Commun. 2025 Jul 1;16(1):5875. doi: 10.1038/s41467-025-61047-y. Nat Commun. 2025. PMID: 40595687 Free PMC article.
Abstract
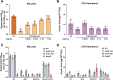
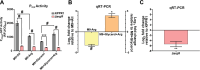
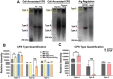
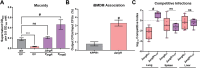
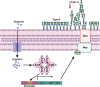
Hypervirulent Klebsiella pneumoniae is associated with severe community-acquired infections. Hypervirulent K. pneumoniae colonies typically exhibit a mucoid phenotype. K. pneumoniae mucoidy is influenced by a complex combination of environmental factors and genetic mechanisms. Mucoidy results from altered capsular polysaccharide chain length, yet the specific environmental cues regulating this phenotype and their impact on pathogenesis remain unclear. This study demonstrates that casamino acids enhance the mucoidy phenotype but do not affect total capsular polysaccharide levels. Through targeted screening of each amino acid present in casamino acids, we identified that arginine is necessary and sufficient to stimulate the mucoid phenotype without altering capsule abundance. Furthermore, arginine activates the rmpADC promoter, increasing rmpD transcript levels, which in turn modulates capsular polysaccharide chain length and diversity. The arginine regulator, ArgR, plays a pivotal role in this regulatory cascade since deleting argR decreases mucoidy and increases capsular polysaccharide chain length diversity. Additionally, the ∆argR mutant displays increased macrophage association and has a substantial competitive defect in the lungs of mice, suggesting a link between arginine-dependent gene regulation, immune evasion and in vivo fitness. We discovered that arginine-dependent regulation of mucoidy is conserved in four additional hypervirulent K. pneumoniae isolates likely via a conserved ARG binding box present in rmp promoters. Our findings support a model in which arginine activates ArgR and increases mucoidy in hypervirulent K. pneumoniae. As a result, it is possible that arginine-dependent regulation of mucoidy allows hypervirulent K. pneumoniae to adapt the cell surface across different niches. This study underscores the significance of arginine as a regulatory signal in bacterial virulence.
Keywords: Klebsiella pneumoniae; amino acids; arginine; capsular polysaccharide; capsule; hypermucoviscosity; hypervirulence; mucoidy.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
