This is a preprint.
Structural impact of 3-methylcytosine modification on the anticodon stem of a neuronally-enriched arginine tRNA
- PMID: 39605410
- PMCID: PMC11601484
- DOI: 10.1101/2024.11.18.624017
Structural impact of 3-methylcytosine modification on the anticodon stem of a neuronally-enriched arginine tRNA
Update in
-
Structural Impact of 3-methylcytosine Modification on the Anticodon Stem-loop of a Neuronally-enriched Arginine tRNA.J Mol Biol. 2025 Aug 15;437(16):169096. doi: 10.1016/j.jmb.2025.169096. Epub 2025 Mar 29. J Mol Biol. 2025. PMID: 40158946
Abstract
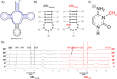
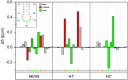
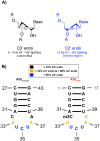
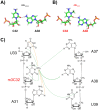
All tRNAs undergo a series of chemical modifications to fold and function correctly. In mammals, the C32 nucleotide in the anticodon loop of tRNA-Arg-CCU and UCU is methylated to form 3-methylcytosine (m3C). Deficiency of m3C in arginine tRNAs has been linked to human neurodevelopmental disorders, indicating a critical biological role for m3C modification. However, the structural repercussions of m3C modification are not well understood. Here, we examine the structural effects of m3C32 modification on the anticodon stem loop (ASL) of human tRNA-Arg-UCU-4-1, a unique tRNA with enriched expression in the central nervous system. Optical melting experiments demonstrate that m3C modification can locally disrupt nearby base pairing within the ASL while simultaneously stabilizing the ASL electrostatically, resulting in little net change thermodynamically. The isoenergetic nature of the C32 - A38 pair vs the m3C32 - A38 pair may help discriminate against structures not adopting canonical C32 - A38 pairings, as most other m3C pairings are unfavorable. Furthermore, multidimensional NMR reveals that after m3C modification there are changes in hairpin loop structure and dynamics, the structure of A37, and the neighboring A31 - U39 base pair. However, these structural changes after modification are made while maintaining the shape of the C32 - A38 pairing, which is essential for efficient tRNA function in translation. These findings suggests that m3C32 modification could alter interactions of tRNA-Arg isodecoders with one or more binding partners while simultaneously maintaining the tRNA's ability to function in translation.
Keywords: 3-methylcytosine; NMR; RNA modification; RNA structure; anticodon stem-loop; m3C; optical melting; tRNA; tRNA-Arg-UCU.
Figures


Similar articles
-
Structural Impact of 3-methylcytosine Modification on the Anticodon Stem-loop of a Neuronally-enriched Arginine tRNA.J Mol Biol. 2025 Aug 15;437(16):169096. doi: 10.1016/j.jmb.2025.169096. Epub 2025 Mar 29. J Mol Biol. 2025. PMID: 40158946
-
DALRD3 encodes a protein mutated in epileptic encephalopathy that targets arginine tRNAs for 3-methylcytosine modification.Nat Commun. 2020 May 19;11(1):2510. doi: 10.1038/s41467-020-16321-6. Nat Commun. 2020. PMID: 32427860 Free PMC article.
-
Evolving specificity of tRNA 3-methyl-cytidine-32 (m3C32) modification: a subset of tRNAsSer requires N6-isopentenylation of A37.RNA. 2016 Sep;22(9):1400-10. doi: 10.1261/rna.056259.116. Epub 2016 Jun 27. RNA. 2016. PMID: 27354703 Free PMC article.
-
The Importance of Being Modified: The Role of RNA Modifications in Translational Fidelity.Enzymes. 2017;41:1-50. doi: 10.1016/bs.enz.2017.03.005. Epub 2017 Apr 22. Enzymes. 2017. PMID: 28601219 Free PMC article. Review.
-
The effect of pseudouridine and pH on the structure and dynamics of the anticodon stem-loop of tRNA(Lys,3).Nucleic Acids Symp Ser. 1997;(36):56-7. Nucleic Acids Symp Ser. 1997. PMID: 9478205 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
