This is a preprint.
MAT2a and AHCY inhibition disrupts antioxidant metabolism and reduces glioblastoma cell survival
- PMID: 39605416
- PMCID: PMC11601785
- DOI: 10.1101/2024.11.23.624981
MAT2a and AHCY inhibition disrupts antioxidant metabolism and reduces glioblastoma cell survival
Update in
-
Methionine cycle inhibition disrupts antioxidant metabolism and reduces glioblastoma cell survival.J Biol Chem. 2025 Apr;301(4):108349. doi: 10.1016/j.jbc.2025.108349. Epub 2025 Feb 25. J Biol Chem. 2025. PMID: 40015640 Free PMC article.
Abstract
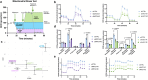
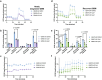
Glioblastoma (GBM) is a highly aggressive primary malignant adult brain tumor that inevitably recurs with a fatal prognosis. This is due in part to metabolic reprogramming that allows tumors to evade treatment. We therefore must uncover the pathways mediating these adaptations to develop novel and effective treatments. We searched for genes that are essential in GBM cells as measured by a whole-genome pan-cancer CRISPR screen available from DepMap and identified the methionine metabolism genes MAT2A and AHCY. We conducted genetic knockdown, evaluated mitochondrial respiration, and performed targeted metabolomics to study the function of these genes in GBM. We demonstrate that MAT2A or AHCY knockdown induces oxidative stress, hinders cellular respiration, and reduces the survival of GBM cells. Furthermore, selective MAT2a or AHCY inhibition reduces GBM cell viability, impairs oxidative metabolism, and changes the metabolic profile of these cells towards oxidative stress and cell death. Mechanistically, MAT2a or AHCY regulates spare respiratory capacity, the redox buffer cystathionine, lipid and amino acid metabolism, and prevents DNA damage in GBM cells. Our results point to the methionine metabolic pathway as a novel vulnerability point in GBM.
Keywords: glioblastoma; lipid peroxidation; metabolism; metabolomics; methionine; mitochondria; oxidative stress.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures




Similar articles
-
Methionine cycle inhibition disrupts antioxidant metabolism and reduces glioblastoma cell survival.J Biol Chem. 2025 Apr;301(4):108349. doi: 10.1016/j.jbc.2025.108349. Epub 2025 Feb 25. J Biol Chem. 2025. PMID: 40015640 Free PMC article.
-
Exosomal circ-AHCY promotes glioblastoma cell growth via Wnt/β-catenin signaling pathway.Ann Clin Transl Neurol. 2023 Jun;10(6):865-878. doi: 10.1002/acn3.51743. Epub 2023 May 7. Ann Clin Transl Neurol. 2023. PMID: 37150844 Free PMC article.
-
Hypoxia-induced one-carbon metabolic reprogramming in glioma stem-like cells.Life Med. 2023 Nov 23;2(6):lnad048. doi: 10.1093/lifemedi/lnad048. eCollection 2023 Dec. Life Med. 2023. PMID: 39872059 Free PMC article.
-
MAT2A inhibition combats metabolic and transcriptional reprogramming in cancer.Drug Discov Today. 2024 Nov;29(11):104189. doi: 10.1016/j.drudis.2024.104189. Epub 2024 Sep 19. Drug Discov Today. 2024. PMID: 39306235 Review.
-
Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis.J Hepatol. 2013 Oct;59(4):830-41. doi: 10.1016/j.jhep.2013.04.031. Epub 2013 May 7. J Hepatol. 2013. PMID: 23665184 Review.
References
-
- Kotecha R., Odia Y., Khosla A. A. & Ahluwalia M. S. (2023) Key Clinical Principles in the Management of Glioblastoma. JCO Oncol. Pract. 19, 180–189 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
