This is a preprint.
Identifying Alzheimer's disease-associated genes using PhenoGeneRanker
- PMID: 39605436
- PMCID: PMC11601490
- DOI: 10.1101/2024.11.12.623269
Identifying Alzheimer's disease-associated genes using PhenoGeneRanker
Abstract
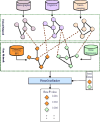
Alzheimer's disease (AD) is a neurogenerative disease that affects millions worldwide with no effective treatment. Several studies have been conducted to decipher to genomic underpinnings of AD. Due to its complex nature, many genes have been found to be associated with AD. Despite these findings, the pathophysiology of the disease is still elusive. To discover new putative AD-associated genes, in this study, we integrated multimodal gene and phenotype datasets of AD using network biology methods to prioritize potential AD-related genes. We constructed a multiplex heterogeneous network composed of patient and gene similarity networks utilizing phenotypic and omics datasets of AD patients from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database. We applied PhenoGeneRanker to traverse this network to discover potential AD-associated genes. To assess the impact of each network layer and seed gene, we also run PhenoGeneRanker on different variants of the network and seed genes. Our results showed that top-ranked genes captured several known AD-related genes and were enriched in Gene Ontology (GO) terms related to AD. We also observed that several top-ranked genes that are not in AD-associated gene list had literature supporting their potential relevance to AD.
Keywords: APP; Alzheimer’s Disease genes; PhenoGeneRanker; gene prioritization; network biology; network propagation.
Conflict of interest statement
Conflicts None declared.
Figures



Similar articles
-
PhenoGeneRanker: Gene and Phenotype Prioritization Using Multiplex Heterogeneous Networks.IEEE/ACM Trans Comput Biol Bioinform. 2022 Sep-Oct;19(5):2950-2962. doi: 10.1109/TCBB.2021.3098278. Epub 2022 Oct 10. IEEE/ACM Trans Comput Biol Bioinform. 2022. PMID: 34283720 Free PMC article.
-
Single-cell network biology characterizes cell type gene regulation for drug repurposing and phenotype prediction in Alzheimer's disease.PLoS Comput Biol. 2022 Jul 18;18(7):e1010287. doi: 10.1371/journal.pcbi.1010287. eCollection 2022 Jul. PLoS Comput Biol. 2022. PMID: 35849618 Free PMC article.
-
An integrated brain-specific network identifies genes associated with neuropathologic and clinical traits of Alzheimer's disease.Brief Bioinform. 2022 Jan 17;23(1):bbab522. doi: 10.1093/bib/bbab522. Brief Bioinform. 2022. PMID: 34953465 Free PMC article.
-
A Review of the Recent Advances in Alzheimer's Disease Research and the Utilization of Network Biology Approaches for Prioritizing Diagnostics and Therapeutics.Diagnostics (Basel). 2022 Nov 28;12(12):2975. doi: 10.3390/diagnostics12122975. Diagnostics (Basel). 2022. PMID: 36552984 Free PMC article. Review.
-
2014 Update of the Alzheimer's Disease Neuroimaging Initiative: A review of papers published since its inception.Alzheimers Dement. 2015 Jun;11(6):e1-120. doi: 10.1016/j.jalz.2014.11.001. Alzheimers Dement. 2015. PMID: 26073027 Free PMC article. Review.
References
-
- Patterson C. (2018). World alzheimer report 2018.
-
- Pudelewicz A., Talarska D. & Bączyk G. Burden of caregivers of patients with Alzheimer’s disease. Scand. J. Caring Sci. 33, 336–341 (2019). - PubMed
-
- Bloom G. S. Amyloid-β and Tau: The Trigger and Bullet in Alzheimer Disease Pathogenesis. JAMA Neurol. 71, 505 (2014). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
