This is a preprint.
Immune Evasion, Cell-Cell Fusion, and Spike Stability of the SARS-CoV-2 XEC Variant: Role of Glycosylation Mutations at the N-terminal Domain
- PMID: 39605558
- PMCID: PMC11601353
- DOI: 10.1101/2024.11.12.623078
Immune Evasion, Cell-Cell Fusion, and Spike Stability of the SARS-CoV-2 XEC Variant: Role of Glycosylation Mutations at the N-terminal Domain
Update in
-
Role of glycosylation mutations at the N-terminal domain of SARS-CoV-2 XEC variant in immune evasion, cell-cell fusion, and spike stability.J Virol. 2025 Apr 15;99(4):e0024225. doi: 10.1128/jvi.00242-25. Epub 2025 Mar 26. J Virol. 2025. PMID: 40135879 Free PMC article.
Abstract
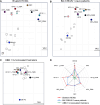
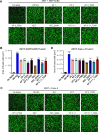
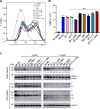
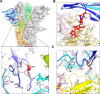
SARS-CoV-2 continues to evolve, producing new variants that drive global COVID-19 surges. XEC, a recombinant of KS.1.1 and KP.3.3, contains T22N and F59S mutations in the spike protein's N-terminal domain (NTD). The T22N mutation, similar to the DelS31 mutation in KP.3.1.1, introduces a potential N-linked glycosylation site in XEC. In this study, we examined the neutralizing antibody (nAb) response and mutation effects in sera from bivalent-vaccinated healthcare workers, BA.2.86/JN.1 wave-infected patients, and XBB.1.5 monovalent-vaccinated hamsters, assessing responses to XEC alongside D614G, JN.1, KP.3, and KP.3.1.1. XEC demonstrated significantly reduced neutralization titers across all cohorts, largely due to the F59S mutation. Notably, removal of glycosylation sites in XEC and KP.3.1.1 substantially restored nAb titers. Antigenic cartography analysis revealed XEC to be more antigenically distinct from its common ancestral BA.2.86/JN.1 compared to KP.3.1.1, with the F59S mutation as a determining factor. Similar to KP.3.1.1, XEC showed reduced cell-cell fusion relative to its parental KP.3, a change attributed to the T22N glycosylation. We also observed reduced S1 shedding for XEC and KP.3.1.1, which was reversed by ablation of T22N and DelS31 glycosylation mutations, respectively. Molecular modeling suggests that T22N and F59S mutations of XEC alters hydrophobic interactions with adjacent spike protein residues, impacting both conformational stability and neutralization. Overall, our findings underscore the pivotal role of NTD mutations in shaping SARS-CoV-2 spike biology and immune escape mechanisms.
Conflict of interest statement
DECLARATION OF INTERESTS The authors have no competing interests to disclose.
Figures


References
-
- Gangavarapu K, Latif AA, Mullen JL, Alkuzweny M, Hufbauer E, Tsueng G, Haag E, Zeller M, Aceves CM, Zaiets K, Cano M, Zhou X, Qian Z, Sattler R, Matteson NL, Levy JI, Lee RTC, Freitas L, Maurer-Stroh S, Core G, Curation T, Suchard MA, Wu C, Su AI, Andersen KG, Hughes LD. 2023. Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat Methods 20:512–522. - PMC - PubMed
-
- Callaway E. 2023. Why a highly mutated coronavirus variant has scientists on alert. Nature 620:934. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
