This is a preprint.
Ethanolamine-induced assembly of microcompartments is required for Fusobacterium nucleatum virulence
- PMID: 39605705
- PMCID: PMC11601286
- DOI: 10.1101/2024.11.11.623001
Ethanolamine-induced assembly of microcompartments is required for Fusobacterium nucleatum virulence
Update in
-
Ethanolamine-induced assembly of microcompartments is required for Fusobacterium nucleatum virulence.mBio. 2025 Feb 5;16(2):e0340524. doi: 10.1128/mbio.03405-24. Epub 2024 Dec 23. mBio. 2025. PMID: 39714188 Free PMC article.
Abstract
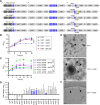
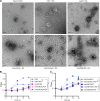
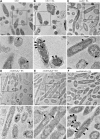
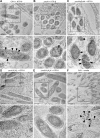
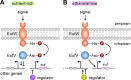
Many bacteria metabolize ethanolamine as a nutrient source through cytoplasmic organelles named bacterial microcompartments (BMCs). Here we investigated the molecular assembly, regulation, and function of BMCs in Fusobacterium nucleatum - a Gram-negative oral pathobiont that is associated with adverse pregnancy outcomes. The F. nucleatum genome harbors a conserved ethanolamine utilization (eut) locus with 21 genes that encode several putative BMC shell proteins and a two-component signal transduction system (TCS), in addition to the enzymes for ethanolamine transport and catabolism. We show that the expression of most of these genes as well as BMC formation is highly increased in wild type fusobacteria when cultured in the presence of ethanolamine as a nutrient source. Deletion of the response regulator EutV eliminated this induction of eut mRNAs and BMCs, thus demonstrating that BMC formation is transcriptionally regulated by the TCS EutV-EutW in response to ethanolamine. Mass spectrometry of isolated BMCs unveiled the identity of the constituent proteins EutL, EutM1, EutM2, and EutN. Consistent with the role of these proteins in BMC assembly and metabolism, deletion of eutN, eutL/eutM 1 /eutM 2 , or eutL/eutM 1 /eutM 2 /eutN not only affected BMC formation, but also ethanolamine utilization, causing cell growth defects with ethanolamine as nutrient. BMCs also assembled in fusobacteria cultured with placental cells or the culture media, a process that is dependent on the BMC shell proteins. Significantly, we show that the eutN mutant is defective in inducing preterm birth in a mouse model. Together, these results establish that BMC-mediated metabolism of ethanolamine is critical for fusobacterial virulence.
Keywords: Fusobacterium nucleatum; bacterial microcompartment; electron microscopy; ethanolamine utilization; preterm birth.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
