Dynamics simulations of hypoxia inducible factor-1 regulatory network in cancer using formal verification techniques
- PMID: 39606028
- PMCID: PMC11599740
- DOI: 10.3389/fmolb.2024.1386930
Dynamics simulations of hypoxia inducible factor-1 regulatory network in cancer using formal verification techniques
Abstract
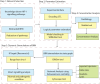
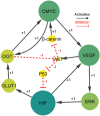
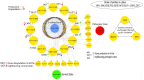
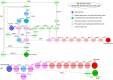
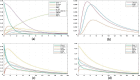
Hypoxia-inducible factor-1 (HIF-1) regulates cell growth, protein translation, metabolic pathways and therefore, has been advocated as a promising biological target for the therapeutic interventions against cancer. In general, hyperactivation of HIF-1 in cancer has been associated with increases in the expression of glucose transporter type-1 (GLUT-1) thus, enhancing glucose consumption and hyperactivating metabolic pathways. The collective behavior of GLUT-1 along with previously known key players AKT, OGT, and VEGF is not fully characterized and lacks clarity of how glucose uptake through this pathway (HIF-1) probes the cancer progression. This study uses a Rene Thomas qualitative modeling framework to comprehend the signaling dynamics of HIF-1 and its interlinked proteins, including VEGF, ERK, AKT, GLUT-1, β-catenin, C-MYC, OGT, and p53 to elucidate the regulatory mechanistic of HIF-1 in cancer. Our dynamic model reveals that continuous activation of p53, β-catenin, and AKT in cyclic conditions, leads to oscillations representing homeostasis or a stable recovery state. Any deviation from this cycle results in a cancerous or pathogenic state. The model shows that overexpression of VEGF activates ERK and GLUT-1, leads to more aggressive tumor growth in a cancerous state. Moreover, it is observed that collective modulation of VEGF, ERK, and β-catenin is required for therapeutic intervention because these genes enhance the expression of GLUT-1 and play a significant role in cancer progression and angiogenesis. Additionally, SimBiology simulation unveils dynamic molecular interactions, emphasizing the need for targeted therapeutics to effectively regulate VEGF and ERK concentrations to modulate cancer cell proliferation.
Keywords: cellular myelocytomatosis oncogene (C-MYC); extracellular single regulated kinase (ERK); glucose transporter-1 (GLUT-1); hypoxia-inducible factor-1 (HIF-1); oglycosylation transferase (OGT); vascular endothelial growth factor (VEGF).
Copyright © 2024 Azhar, Saeed and Jabeen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Baltazar P., Chadha R., Mateus P. (2011). Quantum computation tree logic — model checking and complete calculus. Int. J. Quantum Inf. 6 (2), 219–236. 10.1142/S0219749908003530 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

