Mechanism of a novel Bacillus subtilis JNF2 in suppressing Fusarium oxysporum f. sp. cucumerium and enhancing cucumber growth
- PMID: 39606119
- PMCID: PMC11599245
- DOI: 10.3389/fmicb.2024.1459906
Mechanism of a novel Bacillus subtilis JNF2 in suppressing Fusarium oxysporum f. sp. cucumerium and enhancing cucumber growth
Abstract
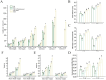
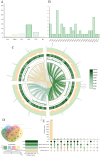
Cucumber Fusarium wilt caused by Fusarium oxysporum f. sp. cucumerium (FOC), is a prevalent soil-borne disease. In this study, Bacillus subtilis JNF2, isolated from the high incidence area of cucumber Fusarium wilt in Luoyang, demonstrated significant inhibitory effects on FOC and promoted cucumber seedling growth. The biocontrol mechanism of strain JNF2 were elucidated through morphological observation, physiological and biochemical experiments, and whole genome sequence analysis. Pot experiments revealed an 81.33 ± 0.21% control efficacy against Fusarium wilt, surpassing the 64.10 ± 0.06% efficacy of hymexazol. Seedlings inoculated with JNF2 exhibited enhanced stem thickness and leaf area compared to control and hymexazol-treated plants. Physiological tests confirmed JNF2's production of indole-3-acetic acid (IAA), siderophores, and hydrolytic enzymes, such as β-1,3-glucanase, amylase, and protease, which inhibited FOC growth and promoted plant development. Genome analysis identified genes encoding antimicrobial peptides and hydrolases, as well as a novel glycocin synthetic gene cluster. These findings underscore B. subtilis JNF2's potential as a biocontrol agent for sustainable cucumber cultivation.
Keywords: Bacillus subtilis; Fusarium oxysporum f. sp. cucumerinum; biocontrol bacteria; growth promotion; whole genome sequence.
Copyright © 2024 Yang, Wang, Jiang, Yao, Liang, Chen, Shi, Tian, Hegazy and Ding.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Evaluation and Genome Analysis of Bacillus subtilis YB-04 as a Potential Biocontrol Agent Against Fusarium Wilt and Growth Promotion Agent of Cucumber.Front Microbiol. 2022 Jun 9;13:885430. doi: 10.3389/fmicb.2022.885430. eCollection 2022. Front Microbiol. 2022. PMID: 35756052 Free PMC article.
-
Genome sequencing and analysis of Bacillus velezensis VJH504 reveal biocontrol mechanism against cucumber Fusarium wilt.Front Microbiol. 2023 Oct 12;14:1279695. doi: 10.3389/fmicb.2023.1279695. eCollection 2023. Front Microbiol. 2023. PMID: 37901818 Free PMC article.
-
Influence of Bacillus subtilis strain Z-14 on microbial ecology of cucumber rhizospheric vermiculite infested with fusarium oxysporum f. sp. cucumerinum.Pestic Biochem Physiol. 2024 May;201:105875. doi: 10.1016/j.pestbp.2024.105875. Epub 2024 Mar 18. Pestic Biochem Physiol. 2024. PMID: 38685217
-
Metabolomic and Transcriptome Analysis of the Inhibitory Effects of Bacillus subtilis Strain Z-14 against Fusarium oxysporum Causing Vascular Wilt Diseases in Cucumber.J Agric Food Chem. 2023 Feb 8;71(5):2644-2657. doi: 10.1021/acs.jafc.2c07539. Epub 2023 Jan 27. J Agric Food Chem. 2023. PMID: 36706360
-
Bacterial endophytome-mediated resistance in banana for the management of Fusarium wilt.3 Biotech. 2021 Jun;11(6):267. doi: 10.1007/s13205-021-02833-5. Epub 2021 May 15. 3 Biotech. 2021. PMID: 34017673 Free PMC article. Review.
Cited by
-
Bacillus subtilis B579 Controls Cucumber Fusarium Wilt by Improving Rhizosphere Microbial Community.Microorganisms. 2025 Jun 13;13(6):1382. doi: 10.3390/microorganisms13061382. Microorganisms. 2025. PMID: 40572270 Free PMC article.
-
Formation of a Novel Antagonistic Bacterial Combination to Enhance Biocontrol for Cucumber Fusarium Wilt.Microorganisms. 2025 Jan 10;13(1):133. doi: 10.3390/microorganisms13010133. Microorganisms. 2025. PMID: 39858901 Free PMC article.
-
Bacillus subtilis ED24 Controls Fusarium culmorum in Wheat Through Bioactive Metabolite Secretion and Modulation of Rhizosphere Microbiome.Microb Ecol. 2025 Aug 19;88(1):89. doi: 10.1007/s00248-025-02590-5. Microb Ecol. 2025. PMID: 40830705 Free PMC article.
References
LinkOut - more resources
Full Text Sources

