Genome sequences of Arthrobacter globiformis phages JuneStar and Pumpkins in Bismarck, ND
- PMID: 39606149
- PMCID: PMC11600132
- DOI: 10.17912/micropub.biology.001378
Genome sequences of Arthrobacter globiformis phages JuneStar and Pumpkins in Bismarck, ND
Abstract
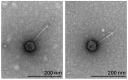
We report the isolation and characteristics of phages JuneStar and Pumpkins, siphoviruses isolated from soil in Bismarck, ND using Arthrobacter globiformis B2979-SEA. Based on gene content similarity, both phages are assigned to actinobacteriophage cluster AZ1. They encode a putative serine integrase that is conserved across cluster AZ1 phages, suggesting a temperate lifestyle.
Copyright: © 2024 by the authors.
Conflict of interest statement
The authors declare that there are no conflicts of interest present.
Figures
References
-
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of molecular biology , 215 (3), 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2 - PubMed
-
- Besemer, J., & Borodovsky, M. (2005). GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic acids research , 33 (Web Server issue), W451–W454. https://doi.org/10.1093/nar/gki487 - PMC - PubMed
-
- Cresawn, S. G., Bogel, M., Day, N., Jacobs-Sera, D., Hendrix, R. W., & Hatfull, G. F. (2011). Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC bioinformatics , 12 , 395. https://doi.org/10.1186/1471-2105-12-395 - PMC - PubMed
-
- Delcher, A. L., Bratke, K. A., Powers, E. C., & Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics (Oxford, England) , 23 (6), 673–679. https://doi.org/10.1093/bioinformatics/btm009 - PMC - PubMed
-
- Hatfull G. F. (2022). Mycobacteriophages: From Petri dish to patient. PLoS pathogens , 18 (7), e1010602. https://doi.org/10.1371/journal.ppat.1010602 - PMC - PubMed
LinkOut - more resources
Full Text Sources
