Autoimmune disease: a view of epigenetics and therapeutic targeting
- PMID: 39606248
- PMCID: PMC11599216
- DOI: 10.3389/fimmu.2024.1482728
Autoimmune disease: a view of epigenetics and therapeutic targeting
Abstract
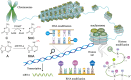
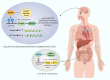
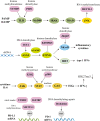
Autoimmune diseases comprise a large group of conditions characterized by a complex pathogenesis and significant heterogeneity in their clinical manifestations. Advances in sequencing technology have revealed that in addition to genetic susceptibility, various epigenetic mechanisms including DNA methylation and histone modification play critical roles in disease development. The emerging field of epigenetics has provided new perspectives on the pathogenesis and development of autoimmune diseases. Aberrant epigenetic modifications can be used as biomarkers for disease diagnosis and prognosis. Exploration of human epigenetic profiles revealed that patients with autoimmune diseases exhibit markedly altered DNA methylation profiles compared with healthy individuals. Targeted cutting-edge epigenetic therapies are emerging. For example, DNA methylation inhibitors can rectify methylation dysregulation and relieve patients. Histone deacetylase inhibitors such as vorinostat can affect chromatin accessibility and further regulate gene expression, and have been used in treating hematological malignancies. Epigenetic therapies have opened new avenues for the precise treatment of autoimmune diseases and offer new opportunities for improved therapeutic outcomes. Our review can aid in comprehensively elucidation of the mechanisms of autoimmune diseases and development of new targeted therapies that ultimately benefit patients with these conditions.
Keywords: DNA methylation; RNA modification; autoimmune disease; epigenetic; histone modification; systemic lupus erythematosus.
Copyright © 2024 Mu, Wang, Liu, Ke, Li, Sun, Zhang and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

