This is a preprint.
Integrative multi-omics profiling of colorectal cancer from a Hispanic/Latino cohort of patients
- PMID: 39606335
- PMCID: PMC11601710
- DOI: 10.1101/2024.11.03.24316599
Integrative multi-omics profiling of colorectal cancer from a Hispanic/Latino cohort of patients
Abstract
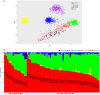
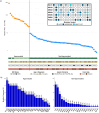
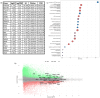
Colorectal cancer contributes to cancer-related deaths and health disparities in the Hispanic and Latino community. To probe both the biological and genetic bases of the disparities, we characterized features of colorectal cancer in terms of somatic alterations and genetic similarity. Specifically, we conducted a comprehensive genome-scale analysis of 67 Hispanic and Latino samples. We performed DNA exome sequencing for somatic mutations, somatic copy number alterations, and genetic similarity. We also performed RNA sequencing for differential gene expression, cellular pathways, and gene fusions. We analyzed all samples for 22 important CRC gene mutations, 8 gene amplifications, and 25 CRC gene fusions. Then, we compared our data from the Hispanic and Latino samples to publicly available, Non-Hispanic White (NHW) cohorts. According to our analyses, twenty-four percent of colorectal carcinomas were hypermutated when patients were of Peruvians-from-Lima-like (1KG-PEL-like) genetic similarity population from the 1000 genome project. Moreover, most of these cases occurred in patients who were less than fiay years old age at diagnosis. Excluding hypermutated tumors, approximately 55% of colon cancers and 58% of rectum cancers exhibited two similar features: 1) the paderns of genomic alterations; 2) percentage of 1KG-PEL-like. We analyzed all samples -- which had a median 1KG-PEL-like proportion of 55% -- for 22 important CRC gene mutations, 8 gene amplifications, and 25 CRC gene fusions. One notable example of a frequently observed gene mutation was SMAD4. Samples with SMAD4 alterations, which are known to support tumor growth and progression, had the highest 1KG-PEL-like proportion (63%). According to our results from risk association analyses and differential gene expression, SMAD4 alterations were significant when we compared Hispanic and Latino samples to NHW cohorts. Of the 8 drug-targetable amplifications, PIK3CA and PI3K exhibited an average 1KG-PEL-like of over 55%. Of the 25 relevant CRC gene fusions, targetable genes included ALK, FGFR1, RAF1, and PTPRK; PTPRK was observed in a sample with the highest 1KG-PEL-like proportion (95%). Using Integrative analysis, we also detected recurrent alterations in the WNT, TGFB, TP53, IGF2/PI3K, and RTK/RAS pathways. Importantly, these alterations mostly occurred in young patients with high 1KG-PEL-like. These findings highlight the potential for tailoring precision medicine therapeutics to an underrepresented population. Our study advances the molecular profiling of CRC in Hispanics and Latinos. In toto, genetic similarity appears to be an important component in understanding colorectal carcinogenesis and has the potential to advance cancer health disparities research.
Figures



References
-
- World Health Organization (2024), International Agency for Research on Cancer. Global Cancer Observatory: Cancer Today. [Accessed on July 04, 2024. Available at: https://gco.iarc.who.int/media/globocan/factsheets/cancers/8-colon-fact-... and https://gco.iarc.who.int/media/globocan/factsheets/cancers/9-rectum-fact...]
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
