This is a preprint.
The ClinGen Syndromic Disorders Gene Curation Expert Panel: Assessing the Clinical Validity of 111 Gene-Disease Relationships
- PMID: 39606380
- PMCID: PMC11601709
- DOI: 10.1101/2024.11.19.24317561
The ClinGen Syndromic Disorders Gene Curation Expert Panel: Assessing the Clinical Validity of 111 Gene-Disease Relationships
Update in
-
The ClinGen Syndromic Disorders Gene Curation Expert Panel: Assessing the clinical validity of 111 gene-disease relationships.Genet Med Open. 2025 Apr 9;3:103429. doi: 10.1016/j.gimo.2025.103429. eCollection 2025. Genet Med Open. 2025. PMID: 40496713 Free PMC article.
Abstract
Purpose: The Clinical Genome Resource (ClinGen) Gene Curation Expert Panels (GCEPs) have historically focused on specific organ systems or phenotypes; thus, the ClinGen Syndromic Disorders GCEP (SD-GCEP) was formed to address an unmet need.
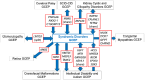
Methods: The SD-GCEP applied ClinGen's framework to evaluate the clinical validity of genes associated with rare syndromic disorders. 111 Gene-Disease Relationships (GDRs) associated with 100 genes spanning the clinical spectrum of syndromic disorders were curated.
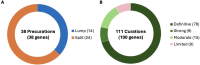
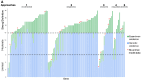
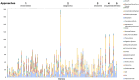
Results: From April 2020 through March 2024, 38 precurations were performed on genes with multiple disease relationships and were reviewed to determine if the disorders were part of a spectrum or distinct entities. 14 genes were lumped into a single disease entity and 24 were split into separate entities, of which 11 were curated by the SD-GCEP. A full review of 111 GDRs for 100 genes followed, with 78 classified as Definitive, 9 as Strong, 15 as Moderate, and 9 as Limited highlighting where further data are needed. All diseases involved two or more organ systems, while the majority (88/111 GDRs, 79.2%) had five or more organ systems affected.
Conclusion: The SD-GCEP addresses a critical gap in gene curation efforts, enabling inclusion of genes for syndromic disorders in clinical testing and contributing to keeping pace with the rapid discovery of new genetic syndromes.
Conflict of interest statement
Conflict of Interest K.B., M.P.B., B.T.B., N.J.B., An.C., Ad.C., A.R.C, K.L.G., A.K., A.M., R.R., Z.Sc., J.P.T., A.J.C. are current or former employees and shareholders of Illumina Inc. K.B., J.M.H., D.L.T., B.W. are employees of Ambry Genetics. K.L.G. is an employee of Rady Children’s Institute for Genomic Medicine. SSJ is the co-founder of Global Gene Corporation Pte Ltd. J.P.T is an employee of Blueprint Genetics (a Quest company). AODL was a paid consultant for Tome Biosciences, Ono Pharma USA, and Addition Therapeutics and receives research funding from Pacific Biosciences. All other authors declare no conflicts of interest.
Figures


References
-
- Christianson Arnold, Howson Christopher P., Modell Bernadette. March of Dimes Global Report on Birth Defects. March of Dimes Birth Defects Foundation; 2006. https://www.prevencioncongenitas.org/wp-content/uploads/2017/02/Global-r...
-
- Mathews TJ, Driscoll AK. Trends in infant mortality in the United States, 2005–2014. NCHS Data Brief. 2017;(279):1–8. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
