This is a preprint.
An orally available Mpro/TMPRSS2 bispecific inhibitor with potent anti-coronavirus efficacy in vivo
- PMID: 39606435
- PMCID: PMC11601862
- DOI: 10.21203/rs.3.rs-5454588/v1
An orally available Mpro/TMPRSS2 bispecific inhibitor with potent anti-coronavirus efficacy in vivo
Update in
-
An orally available Mpro/TMPRSS2 bispecific inhibitor with potent anti-coronavirus efficacy in vivo.Nat Commun. 2025 Jul 16;16(1):6541. doi: 10.1038/s41467-025-60832-z. Nat Commun. 2025. PMID: 40670362 Free PMC article.
Abstract
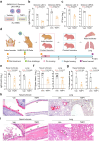
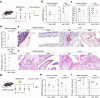
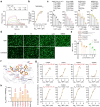
Coronaviruses have caused three major endemics in the past two decades. Alarmingly, recent identification of novel zoonotic coronaviruses that caused human infections suggests the risk of future coronavirus outbreak caused by spillover infection from animal reservoirs remains high1,2. Therefore, development of novel therapeutic options with broad-spectrum anti-coronavirus activities are urgently needed. Here, we develop an orally-available bispecific inhibitor, TMP1, which simultaneously targets key coronavirus replication protease Mpro and the essential airway protease TMPRSS23,4. TMP1 shows broad-spectrum protection not only against different SARS-CoV-2 variants but also against multiple human-pathogenic coronaviruses in vitro. By using the K18-hACE2 transgenic mouse, hDPP4 knock-in mouse and golden Syrian hamster models, we demonstrate TMP1 cross-protects against highly-pathogenic coronaviruses (SARS-CoV-1, SARS-CoV-2 and MERS-CoV) in vivo and efficiently abrogates SARS-CoV-2 transmission. Through structural and mutagenesis studies, we confirmed the direct interaction of TMP1 with Mpro and TMPRSS2, and pinpoint the key sites of interactions. Importantly, TMP1 inhibits the infection of nirmatrelvir-resistant SARS-CoV-2 escape mutants. Together, our findings demonstrate the antiviral potential of the novel bispecific Mpro/TMPRSS2 antiviral design against human-pathogenic coronaviruses and other emerging coronaviruses.
Keywords: COVID-19; Mpro; SARS-CoV-2; TMPRSS2; bispecific inhibitor; in vivo.
Figures



Similar articles
-
An orally available Mpro/TMPRSS2 bispecific inhibitor with potent anti-coronavirus efficacy in vivo.Nat Commun. 2025 Jul 16;16(1):6541. doi: 10.1038/s41467-025-60832-z. Nat Commun. 2025. PMID: 40670362 Free PMC article.
-
Potent 3CLpro inhibitors effective against SARS-CoV-2 and MERS-CoV in animal models by therapeutic treatment.mBio. 2024 Feb 14;15(2):e0287823. doi: 10.1128/mbio.02878-23. Epub 2023 Dec 21. mBio. 2024. PMID: 38126789 Free PMC article.
-
Evaluation of in vitro antiviral activity of SARS-CoV-2 Mpro inhibitor pomotrelvir and cross-resistance to nirmatrelvir resistance substitutions.Antimicrob Agents Chemother. 2023 Nov 15;67(11):e0084023. doi: 10.1128/aac.00840-23. Epub 2023 Oct 6. Antimicrob Agents Chemother. 2023. PMID: 37800975 Free PMC article.
-
Non-pharmacological measures implemented in the setting of long-term care facilities to prevent SARS-CoV-2 infections and their consequences: a rapid review.Cochrane Database Syst Rev. 2021 Sep 15;9(9):CD015085. doi: 10.1002/14651858.CD015085.pub2. Cochrane Database Syst Rev. 2021. PMID: 34523727 Free PMC article.
-
SARS-CoV-2-neutralising monoclonal antibodies for treatment of COVID-19.Cochrane Database Syst Rev. 2021 Sep 2;9(9):CD013825. doi: 10.1002/14651858.CD013825.pub2. Cochrane Database Syst Rev. 2021. PMID: 34473343 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

