This is a preprint.
Evolutionary loss of an antibiotic efflux pump increases Pseudomonas aeruginosa quorum sensing mediated virulence in vivo
- PMID: 39606469
- PMCID: PMC11601840
- DOI: 10.21203/rs.3.rs-5391023/v1
Evolutionary loss of an antibiotic efflux pump increases Pseudomonas aeruginosa quorum sensing mediated virulence in vivo
Update in
-
Evolutionary loss of an antibiotic efflux pump increases Pseudomonas aeruginosa quorum sensing mediated virulence in vivo.Nat Commun. 2025 Sep 25;16(1):8397. doi: 10.1038/s41467-025-63284-7. Nat Commun. 2025. PMID: 40998772 Free PMC article.
Abstract
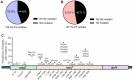
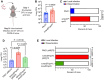
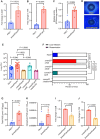
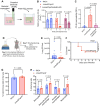
Antibiotic resistance is one of the most pressing threats to human health, yet recent work highlights how loss of resistance may also drive pathogenesis in some bacteria. In two recent studies, we found that β-lactam antibiotic and nutrient stresses faced during infection selected for the genetic inactivation of the Pseudomonas aeruginosa (Pa) antibiotic efflux pump mexEFoprN. Unexpectedly, efflux pump mutations increased Pa virulence during infection; however, neither the prevalence of efflux pump inactivating mutations in real human infections, nor the mechanisms driving increased virulence of efflux pump mutants are known. We hypothesized that human infection would select for efflux pump mutations that drive increased virulence in Pa clinical isolates. Using genome sequencing of hundreds of Pa clinical isolates, we show that mexEFoprN efflux pump inactivating mutations are enriched in Pa cystic fibrosis isolates relative to Pa intensive care unit clinical isolates. Combining RNA-seq, metabolomics, genetic approaches, and infection models we show that efflux pump mutants have elevated expression of two key Pa virulence factors, elastase and rhamnolipids, which increased Pa virulence and lung damage during both acute and chronic infections. Increased virulence factor production was driven by higher Pseudomonas quinolone signal levels in the efflux pump mutants. Finally, genetic restoration of the efflux pump in a representative ICU clinical isolate and the notorious CF Pa Liverpool epidemic strain reduced their virulence. Together, our findings suggest that mutations inactivating antibiotic resistance mechanisms could lead to greater patient mortality and morbidity.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest.
Figures
References
-
- Carmeli Y., Troillet N., Karchmer a W., and Samore M.H. (1999). Health and economic outcomes of antibiotic resistance in Pseudomonas aeruginosa. Archives of internal medicine 159, 1127–1132. - PubMed
-
- Nathwani D., Raman G., Sulham K., Gavaghan M., and Menon V. (2014). Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control 3, 32. 10.1186/2047-2994-3-32. - DOI - PMC - PubMed
-
- Jorth P., McLean K., Ratjen A., Secor P.R., Bautista G.E., Ravishankar S., Rezayat A., Garudathri J., Harrison J.J., Harwood R.A., et al. (2017). Evolved Aztreonam Resistance Is Multifactorial and Can Produce Hypervirulence in Pseudomonas aeruginosa. mBio 8, e00517–17. 10.1128/mBio.00517-17. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

