This is a preprint.
ATP-dependent thermoring basis for the heat unfolding of the first nucleotide-binding domain isolated from human CFTR
- PMID: 39606474
- PMCID: PMC11601864
- DOI: 10.21203/rs.3.rs-5479740/v1
ATP-dependent thermoring basis for the heat unfolding of the first nucleotide-binding domain isolated from human CFTR
Abstract
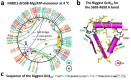
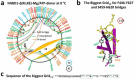
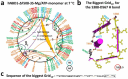
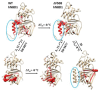
Traditionally, the thermostability of a protein is defined by a melting temperature, at which half of the protein is unfolded. However, this definition cannot indicate the structural origin of a heat-induced unfolding pathway. Here, the thermoring structures were studied on the ATP-dependent heat-induced unfolding of the first nucleotide-binding domain from the human cystic fibrosis transmembrane conductance regulator. The results showed that initial theoretical and experimental melting thresholds aligned well after three structural perturbations including the F508del mutation, the most common cause of cystic fibrosis. This alignment further demonstrated that the heat-induced unfolding process began with the disruption of the least-stable noncovalent interaction within the biggest thermoring along the single peptide chain. The C-terminal region, which was related to the least-stable noncovalent interaction and the ATP-dependent dimerization of two nucleotide-binding domains, emerged as a crucial determinant of the thermal stability of the isolated protein and a potential interfacial drug target to alleviate the thermal defect caused by the F508del mutation. This groundbreaking discovery significantly advances our understanding of protein activity, thermal stability, and molecular pathology.
Keywords: cooperative misfolding; grid thermodynamic signature; least-stable interaction; melting threshold; noncovalent structure; partial unfolding.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT The author declares no conflict of interest. Additional Declarations: The authors declare no competing interests.
Figures


References
-
- Ingram V.M. Gene mutations in human haemoglobin:thechemical difference between normal and sickle cell haemoglobin. Nature 180, 326–328 (1957). - PubMed
-
- Malay AD, Procious SL, Tolan DR. The temperature dependence of activity and structure for the most prevalent mutant aldolase B associated with hereditary fructose intolerance. Arch Biochem Biophys.408, 295–304 (2002). - PubMed
-
- Treweek TM, Rekas A, Lindner RA, Walker MJ, Aquilina JA, Robinson CV, Horwitz J, Perng MD, Quinlan RA, Carver JA. R120G alphaB-crystallin promotes the unfolding of reduced alpha-lactalbumin and is inherently unstable. FEBS J.272, 711–24 (2005). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

