This is a preprint.
WelQrate: Defining the Gold Standard in Small Molecule Drug Discovery Benchmarking
- PMID: 39606732
- PMCID: PMC11601797
WelQrate: Defining the Gold Standard in Small Molecule Drug Discovery Benchmarking
Abstract
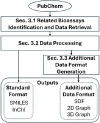
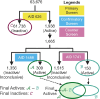
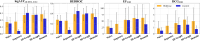
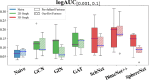
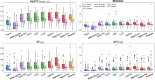
While deep learning has revolutionized computer-aided drug discovery, the AI community has predominantly focused on model innovation and placed less emphasis on establishing best benchmarking practices. We posit that without a sound model evaluation framework, the AI community's efforts cannot reach their full potential, thereby slowing the progress and transfer of innovation into real-world drug discovery. Thus, in this paper, we seek to establish a new gold standard for small molecule drug discovery benchmarking, WelQrate. Specifically, our contributions are threefold: WelQrate Dataset Collection - we introduce a meticulously curated collection of 9 datasets spanning 5 therapeutic target classes. Our hierarchical curation pipelines, designed by drug discovery experts, go beyond the primary high-throughput screen by leveraging additional confirmatory and counter screens along with rigorous domain-driven preprocessing, such as Pan-Assay Interference Compounds (PAINS) filtering, to ensure the high-quality data in the datasets; WelQrate Evaluation Framework - we propose a standardized model evaluation framework considering high-quality datasets, featurization, 3D conformation generation, evaluation metrics, and data splits, which provides a reliable benchmarking for drug discovery experts conducting real-world virtual screening; Benchmarking - we evaluate model performance through various research questions using the WelQrate dataset collection, exploring the effects of different models, dataset quality, featurization methods, and data splitting strategies on the results. In summary, we recommend adopting our proposed WelQrate as the gold standard in small molecule drug discovery benchmarking. The WelQrate dataset collection, along with the curation codes, and experimental scripts are all publicly available at WelQrate.org.
Figures

References
-
- Wognum Cas, Ash Jeremy R, Aldeghi Matteo, Rodríguez-Pérez Raquel, Fang Cheng, Cheng Alan C, Price Daniel J, Clevert Djork-Arné, Engkvist Ola, and Walters W Patrick. A call for an industry-led initiative to critically assess machine learning for real-world drug discovery. Nature Machine Intelligence, pages 1–2, 2024.
-
- Walters Pat. We Need Better Benchmarks for Machine Learning in Drug Discovery. https://practicalcheminformatics.blogspot.com/2023/08/we-need-better-ben..., 2023. [Online; accessed 05-June-2024].
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
