Leveraging Dual-Ligase Recruitment to Enhance Protein Degradation via a Heterotrivalent Proteolysis Targeting Chimera
- PMID: 39606859
- PMCID: PMC11638965
- DOI: 10.1021/jacs.4c11556
Leveraging Dual-Ligase Recruitment to Enhance Protein Degradation via a Heterotrivalent Proteolysis Targeting Chimera
Abstract
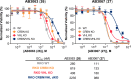
Proteolysis targeting chimera (PROTAC) degraders are typically bifunctional with one E3 ligase ligand connected to one target protein ligand via a linker. While augmented valency has been shown with trivalent PROTACs targeting two binding sites within a given target protein, or used to recruit two different targets, the possibility of recruiting two different E3 ligases within the same compound has not been demonstrated. Here we present dual-ligase recruitment as a strategy to enhance targeted protein degradation. We designed heterotrivalent PROTACs composed of CRBN, VHL and BET targeting ligands, separately tethered via a branched trifunctional linker. Structure-activity relationships of 12 analogues qualifies AB3067 as the most potent and fastest degrader of BET proteins, with minimal E3 ligase cross-degradation. Comparative kinetic analyses in wild-type and ligase single and double knockout cell lines revealed that protein ubiquitination and degradation induced by AB3067 was contributed to by both CRBN and VHL in an additive fashion. We further expand the scope of the dual-ligase approach by developing a heterotrivalent CRBN/VHL-based BromoTag degrader and a tetravalent PROTAC comprising of two BET ligand moieties. In summary, we provide proof-of-concept for dual-E3 ligase recruitment as a strategy to boost degradation fitness by recruiting two E3 ligases with a single degrader molecule. This approach could potentially delay the outset of resistance mechanisms involving loss of E3 ligase functionality.
Conflict of interest statement
The authors declare the following competing financial interest(s): The University of Dundee has filed a PCT patent application PCT/GB2024/051314 on May 24, 2024 covering the chemical structures and their use. A.C., A.G.B., and N. M. are inventors of this patent. A.C. is a scientific founder and shareholder of Amphista Therapeutics, a company that is developing targeted protein degradation therapeutic platforms. The Ciulli laboratory receives or has received sponsored research support from Almirall, Amgen, Amphista Therapeutics, Boehringer Ingelheim, Eisai, Merck KaaG, Nurix Therapeutics, Ono Pharmaceutical and Tocris-Biotechne. G.E.W. is scientific founder and shareholder of Proxygen and Solgate. The Winter lab received research funding from Pfizer. C.M.B., E.A.C., M.U. and K.M.R. are or were employees of Promega Corporation. Promega Corporation is the commercial owner by assignment of patents of the HaloTag, NanoLuc, NanoBRET target engagement, and HiBiT technologies and their applications. Other authors declare no competing interests.
Figures


















References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

