The role of the P2X7 receptor in inactivated SARS-CoV-2-induced lung injury
- PMID: 39607622
- PMCID: PMC12222604
- DOI: 10.1007/s11302-024-10062-7
The role of the P2X7 receptor in inactivated SARS-CoV-2-induced lung injury
Abstract
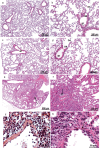
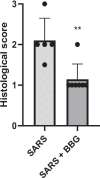
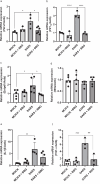
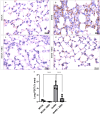
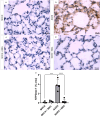
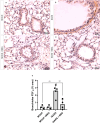
Purinergic signaling plays a role in the pathophysiology of different viral infections. Recently, we showed that COVID-19 increases extracellular ATP levels, which may amplify the pro-inflammatory signals in the disease. The P2X7 receptor can be a protagonist in the pro-inflammatory responses. Herein, we investigated the role of the P2X7 receptor in the lung immune response triggered by inoculation of inactivated SARS-CoV-2 (iSARS-CoV-2) in K18-Human ACE2 transgenic mice. Pharmacological inhibition of the P2X7 receptor was performed with intraperitoneal administration of 50 mg/kg of Brilliant Blue G (BBG) one day before viral inoculation. Animals were divided into four groups: a control group (MOCK), a group inoculated with the inactivated virus iSARS-CoV-2, a BBG-treated control group (MOCK + BBG), and a BBG-treated inoculated group (iSARS-CoV-2 + BBG). Virus inoculation was intratracheal with 50 µl of mock or 2 × 106 Plaque Forming Units (PFU) of iSARS-CoV-2. After three days, blood and lungs were collected. We found a significant increase in ATP and LDH in serum and mRNA levels of P2X7 and P2Y12 receptors, CD39, IL-1β, and TNF-α in the lung of the iSARS-CoV-2 group when compared with the control group. BBG treatment attenuated these increases. Lung histological analyses showed severe lung damage in the iSARS-CoV-2 group, which was reduced by the BBG treatment. Immunohistochemical staining confirmed the increased presence of P2X7, P2Y12, and CD39 proteins in the iSARS-CoV-2 vs. the MOCK group. Thus, P2X7 receptor inhibition decreases iSARS-CoV-2-induced lung inflammation, indicating that this receptor might contribute to SARS-CoV-2 pathology.
Keywords: ATP; CD39; COVID-19; Lung inflammation; P2Y12; Purinergic signaling.
© 2024. The Author(s), under exclusive licence to Springer Nature B.V.
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: The Institutional Animal Care and Use Committee of the Federal University of Rio de Janeiro (protocol number 088/21) reviewed and approved the animal study.
Figures



References
-
- Ruan YJ, Wei CL, Ee AL, Vega VB, Thoreau H, Su ST, Chia JM, Ng P, Chiu KP, Lim L, Zhang T, Peng CK, Lin EO, Lee NM, Yee SL, Ng LF, Chee RE, Stanton LW, Long PM, Liu ET (2003) Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 361:1779–1785 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- SEI-260003/015688/21/Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro - FAPERJ
- E-26/210.240/2020/Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro - FAPERJ
- 406020/2022-1/Instituto Nacional em Ciência e Tecnologia Saúde Cerebral (INCT-Saúde Cerebral/CNPq)
- 03/11/Mercosur Structural Convergence Fund (FOCEM, Mercosur
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

