Amide conjugates of the jasmonate precursor cis-(+)-12-oxo-phytodienoic acid regulate its homeostasis during plant stress responses
- PMID: 39607728
- PMCID: PMC11663710
- DOI: 10.1093/plphys/kiae636
Amide conjugates of the jasmonate precursor cis-(+)-12-oxo-phytodienoic acid regulate its homeostasis during plant stress responses
Abstract
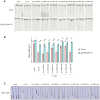
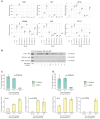
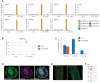
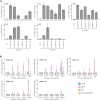
Jasmonates are a family of oxylipin phytohormones regulating plant development and growth and mediating "defense versus growth" responses. The upstream JA biosynthetic precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA) acts independently of CORONATIVE INSENSITIVE 1-mediated JA signaling in several stress-induced and developmental processes. However, its perception and metabolism are only partially understood. An isoleucine analog of the biologically active JA-Ile, OPDA-Ile, was detected years ago in wounded leaves of flowering plants, opening up the possibility that conjugation of cis-OPDA to amino acids might be a relevant mechanism for cis-OPDA regulation. Here, we extended the analysis of amino acid conjugates of cis-OPDA and identified naturally occurring OPDA-Val, OPDA-Phe, OPDA-Ala, OPDA-Glu, and OPDA-Asp accumulating in response to biotic and abiotic stress in Arabidopsis (Arabidopsis thaliana). The OPDA amino acid conjugates displayed cis-OPDA-related plant responses in a JA-Ile-dependent manner. We also showed that the synthesis and hydrolysis of cis-OPDA amino acid conjugates are mediated by members of the amidosynthetase GRETCHEN HAGEN 3 and the amidohydrolase INDOLE-3-ACETYL-LEUCINE RESISTANT 1/ILR1-like families. Thus, OPDA amino acid conjugates function in the catabolism or temporary storage of cis-OPDA in stress responses instead of acting as chemical signals per se.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement: None declared.
Figures

References
-
- Arnold MD, Gruber C, Floková K, Miersch O, Strnad M, Novák O, Wasternack C, Hause B. The recently identified isoleucine conjugate of cis-12-oxo-phytodienoic acid is partially active in cis-12-oxo-phytodienoic acid-specific gene expression of Arabidopsis thaliana. PLoS One. 2016:11(9):e0162829. 10.1371/journal.pone.0162829 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

